Monitoring Microorganisms in the Food Industry
Info: 5597 words (22 pages) Dissertation
Published: 7th Sep 2021
Tagged: Food and Nutrition
Executive Summary
This literature review provides a comprehensive overview of current technologies that are used to monitor microorganisms in the food industry. A focus is placed on monitoring fermenting organisms offline (by taking samples) and online (in situ, and ex situ) by analyzing exiting gas streams and spectrometric monitoring.
Additionally, technologies designated for the detection and monitoring of pathogenic bacteria are discussed. Common technologies and detection methods used in the food industry include indicator organisms, plate counts, Most Probable Number (MPN), dye-reduction tests, immunological analysis, and polymerase chain reaction (PCR).
It is important to note that this literature review does not provide an in-depth analysis of all technologies available, but rather a summary of the most used technologies that are found in industry.
In addition, food and environmental standards were discussed, particularly Hazard Analysis & Critical Control Points (HACCP), Good Manufacturing Processes (GMPs), and Sanitation Standard Operating Procedures (SSOPs).
Although this review is sufficient for the technologies and standards that are currently available in the industry, technologies are constantly evolving to optimize processes and make them the most rapid, specific, and inexpensive as possible.
Table of Contents
2.0 Microorganisms in the food industry
3.0 Monitoring Fermenting Organisms
3.3.1 Exit or Off Gas Analysis
3.3.2 Spectrometric Monitoring
4.0 Methods for Microbiological Examination of foods
4.2 Classical Enumeration Methods
4.5 Polymerase Chain Reaction-Based Methods
6.0 Environmental Monitoring Standards
6.1 Hazard Analysis & Critical Control Points
6.2 Good Manufacturing Practices
6.3 Sanitation Standard Operating Procedures
7.0 Conclusion and Future Prospects
List of Figures
Figure 1: Online Fermentation Monitoring Probes (a) in situ (b) ex situ.
Figure 2: Schematic representation of the methods used to detect foodborne pathogens.
Figure 3: ‘pour’ plate and ‘spread’ plate method
Figure 4: Flowsheet of MPN test for E. coli.
List of Tables
Table 1: European Standards for pathogenic microorganisms in meat.
1.0 Introduction
A wide range of microorganisms from branches of yeast and bacteria are used in the food industry. The primary use of these organisms is for the production of bakery products, dairy products, wine, and beer through fermentation (Batt, 2014). Fermentation is also a helpful process for preserving food products (Batt, 2014). A variety of technologies and methods can be used to monitor these fermenting organisms. Monitoring these processes are essential to ensure product consistency, safety, and optimal economic efficiency (McNeil, Archer, Giavasis, & Harvey, 2013). Fermenting organisms and monitoring techniques will be discussed later in this literature review.
Microorganisms can also be present in the food industry in the form of pathogens (Batt, 2014). Pathogens such as E. coli and Salmonella, which are commonly found in food, can be detrimental to human health if consumed. It is important to remove these pathogens from food and water before they are consumed to ensure the safety and well-being of the consumer. To ensure the quality of food, a variety of tests are used to detect pathogenic organisms on the food and the surrounding environment (Filion, 2012).
In this literature review, common detection tests, including enumeration methods, immunological analysis, and PCR, will be discussed. Government regulations and Canadian Hygiene and Safety Standards for food and the surrounding environment, such as Hazard Analysis & Critical Control Points (HACCP), Good Manufacturing Processes (GMPs), and Sanitation Standard Operating Procedures (SSOPs) will also be reviewed.
2.0 Microorganisms in the food industry
Microorganisms can be found everywhere around us, from the ground we walk on to the air that we breathe. Food is no exception. A wide range of microorganisms can be found in food, some that are beneficial to humans and others that are detrimental to human health. The following sections outline the most common microorganisms found in food.
2.1 Fermenting Organisms
Fermentation is defined as a biotechnical process that uses microorganisms to perform enzyme-catalyzed transformation of organic matter (Sanchez, 2008). These organisms (yeast and bacteria) have biological activities that produce enzymes which break down substrates (Sanchez, 2008). In the food industry, fermentation is used to: (1) develop a wide range of flavors, scents, and textures in food; (2) preserve food using lactic acid, alcohol, acetic acid, and alkaline fermentations; (3) enrich food with essential proteins, amino acids, fatty acids, and vitamins; (4) detoxify fermented products; and (5) reduce cooking time for food (Sanchez, 2008).
2.1.1 Yeasts
Yeasts are fungi that asexually reproduce by budding or fission, which results in growth primarily comprised of single cells (Kurtzman, Fell, & Boekhout, 2011). The primary role of yeast in the food industry is the fermentation of beers, cider, wines, distilled spirits, bakery products, cheese, sausages, and other fermented food (Kurtzman et al., 2011). These products are produced by the breakdown of glucose to ethanol and carbon dioxide. Yeast species such as Saccharomyces cerevisiae, Ogataea polymorpha, and Komatagataella pastoris have also been developed as industrial organisms for the production of proteins and enzymes (Kurtzman et al., 2011). Saccharomyces cerevisiae is the principal yeast used worldwide due to its key role in many food fermentations (Kurtzman et al., 2011).
2.1.2 Lactic Acid Bacteria
Lactic Acid Bacteria (LAB) are nonsporulating gram-positive cocci or rods that produce lactic acids as a fermentation product of carbohydrates (Lahtinen, Ouwehand, Salminen, & Wright, 2011). LAB are particularly used to ferment milk products such as yogurt, cheese, and butter (Lahtinen et al., 2011)
2.2 Mold
A mold is a type of fungus that grows as multicellular filaments called hyphae. Mold is a common food contaminant which, under the correct conditions (cool and damp environments), can reproduce rapidly and cause food spoilage.
2.3 Pathogens
A pathogen is an organism that lives on or in a host organism. Primary pathogens can cause acute, life-threatening infections and can spread rapidly from host to host (Alberts, 2017). Other pathogens infect an individual without causing disease (Alberts, 2017). Pathogens can be divided into two groups: Non-bacterial agents and bacterial agents. Escherichia coli, Salmonella, Cronobacter Species, and Mycobacterium Species are common pathogenic bacteria found in food (Adams, Moss, & McClure, 2016).
These bacteria can cause side effects including, but not limited to, abdominal cramping, vomiting, fatigue, fever, diarrhea, and death, if not treated promptly (Adams et al., 2016). Non-bacterial agents include Helminths and Nematodes, Protozoa, Toxigenic Algae, Toxigenic Fungi, Foodborne Viruses, and Spongiform Encephalopathies (Adams et al., 2016). These agents can also cause debilitating infections if consumed (Adams et al., 2016).
3.0 Monitoring Fermenting Organisms
Before discussing the technologies employed to monitor fermentation, it is important to understand the types of fermentation processes used for food manufacture. There are two distinct types of fermentation processes; the oldest being solid substrate fermentation (SSF), and the other being submerged liquid fermentation (SLF) (McNeil et al., 2013). SSF has lower energy inputs than SLF, simpler reactor systems, suitability for small local scale production, ease of product recovery, and the ability to use waste materials as substrates for growth (McNeil et al., 2013). On the other hand, SSF systems are heterogenous, which presents difficulties in taking representative samples and effective product control throughout the reactor (McNeil et al., 2013). As a result, most fermented foods are now produced using SLF. The technologies discussed in the following section pertain to SLF processes.
3.1 Rationale for Monitoring
Monitoring SLF processes are necessary to ensure product consistency and safety. Additionally, by monitoring SLF, nutrient utilization, yield, and productivity are measured and can be optimized to reduce process economics (McNeil et al., 2013).
3.2 Offline Monitoring
In offline monitoring, a representative sample is taken from the fermenter and analyzed later in a laboratory (McNeil et al., 2013). In large fermenters, there may be inhomogeneity in the mixture which makes gathering a representative sample challenging (McNeil et al., 2013). Another disadvantage to this method is that it does not give real-time data for the conditions in the fermenter (McNeil et al., 2013). On the other hand, if the sample is stored at suitable conditions (chilling or freezing with preservative agents), offline analysis such as nuclear magnetic resonance (NMR), gas liquid chromatography (GLC), and high-performance liquid chromatography (HPLC) is very effective in analyzing large numbers of samples (McNeil et al., 2013). As a result, routine analysis for most fermenters is carried out offline (McNeil et al., 2013).
3.3 Online Monitoring
Real-time information can be attained by in situ or ex situ sensors as shown in Figure 1 below (Vojinović, Cabral, & Fonseca, 2006). Ex situ sampling is where the sample is withdrawn from the bulk of the material and transported to a testing area (Vojinović et al., 2006). In situ sampling is where the test is conducted directly in the bulk material (Vojinović et al., 2006). 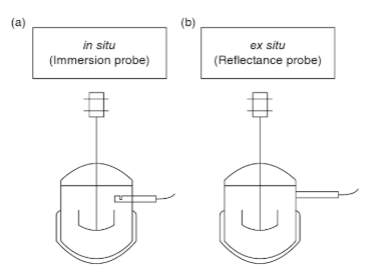
Figure 1: Online Fermentation Monitoring Probes (a) in situ (b) ex situ.
The ideal sensor for food manufacturing should be reliable, inexpensive, physically resilient, simple to implement and operate, capable of generating real-time information, and capable of monitoring multiple fermenters at a time (McNeil et al., 2013).
3.3.1 Exit or Off Gas Analysis
Most fermentations can be readily monitored ex situ using off gas analysis (McNeil et al., 2013). Many fermentation processes are aerobic, where sterile air is supplied to the system for intake by the fermenting bacteria (McNeil et al., 2013). In this process, oxygen uptake and CO2 evolution are stoichiometrically linked, which can be used to evaluate the growth of microorganisms (McNeil et al., 2013).
From the real-time gas values of CO2, it is possible to determine the oxygen uptake rate (OUR), carbon dioxide evolution rate (CER), and the respiratory quotient (RQ) (McNeil et al., 2013). It is then straightforward to generate models that accurately depict the specific growth rate and substrate consumption of the cultures (McNeil et al., 2013).
Since the sensors sit on the outside of the fermenting environment, they do not need to be as physically tough as in situ sensors. As a result, these sensors are commonly less expensive. Conversely, this technique can not be used to account for the mass balance of elements in the fermenter. For instance, it cannot account for all the carbon and energy that has been used by a culture (McNeil et al., 2013).
3.3.2 Spectrometric Monitoring
Most chemical, enzyme, or electrochemical measurement techniques have been adapted to in situ operations in fermenters (McNeil et al., 2013). For instance, sensors for temperature, pH, and pO2 are used routinely to monitor bioprocesses in laboratories (Vojinović et al., 2006). Enzyme-based sensors have also been developed for monitoring fermentation analytes such as glucose.
In this method, a selectively permeable layer is used to trap suitable enzymes (McNeil et al., 2013). When an analyte such as glucose diffuses through the membrane, it reacts with an enzyme (McNeil et al., 2013). The probe then emits a signal output that is proportionate to the extent of the reaction, which can be tracked to monitor the fermentation (McNeil et al., 2013). Although this method has been widely investigated, these sensors are of little use at production scale in food fermentations due to the increased risk to process sterility (McNeil et al., 2013). Spectrophotometric methods have proven to be very helpful in situ food process monitoring. Some common forms of spectroscopy are outlined below.
3.3.2.1 Near-infrared Spectroscopy (NIRS)
The near-infrared region occupies the wavelength range from 700-2500 nm in the electromagnetic spectrum (McNeil et al., 2013). Most absorbances in this region arise from X-H bonds (C-H, 0H, etc.) (McNeil et al., 2013). Since these bonds are found in abundance in biological systems, NIR provides information regarding most analytes in a fermentation system (McNeil et al., 2013).
NIR spectroscopy is fast, multi-analyte, non-destructive, non-invasive, and can be implemented in situ using a stainless steel fiber optic probe system (McNeil et al., 2013). Additionally, the relatively weak absorbances in the NIR region makes it ideal to analyze samples that are highly light scattering and light absorbing, such as that of fermentation fluid (McNeil et al., 2013). On the other hand, the weak NIR region makes it difficult to analyze low analyte concentrations (McNeil et al., 2013).
3.3.2.2 Mid-infrared Spectroscopy (MIRS)
Due to stronger absorbances in the fundamental IR, lower analyte levels can be measurable using MIRS (McNeil et al., 2013). Although studies have proven the potential for MIRS in food fermentation, MIRS has been less employed than NIRS. In comparison to NIRS, MIRS is more costly because the probes require expensive wave guides and multiplexing technology (McNeil et al., 2013).
3.3.2.3 Raman Spectroscopy
Raman spectroscopy is used even less than MIRS in fermentation monitoring due to the high cost of Raman systems in comparison to NIRS/MIRS (McNeil et al., 2013). Although the results from Raman spectroscopy are highly accurate, due to the cost and complexity of the Raman system, it will be a long time before this system is used as a routine tool (McNeil et al., 2013).
4.0 Methods for Microbiological Examination of foods
In the previous section, positive uses for microorganisms are outlined. Although some microorganisms may be beneficial to humans, most organisms found in food are harmful to human health. Foodborne pathogens are the cause of many diseases worldwide (Forsythe, 2012). It is important to test and monitor food production to ensure:
- the food meets standard regulations (will be discussed later);
- food meets internal standards set by the processing company or purchaser;
- food materials entering the factory meet standards from the supplier; and
- sanitation in the surrounding environment is being maintained (Forsythe, 2012).
Figure 2 outlines the detection methods used in the food industry for the detection of foodborne pathogens (Priyanka, Patil, & Dwarakanath, 2016). Selected methods will be discussed further. 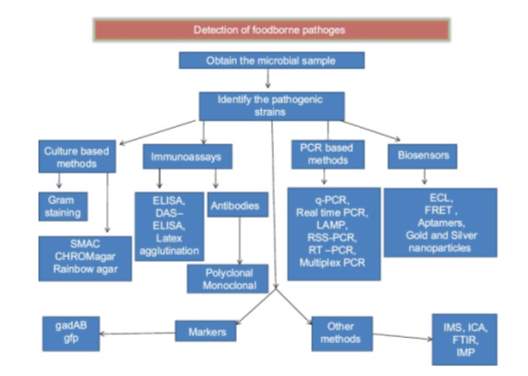
Figure 2: Schematic representation of the methods used to detect foodborne pathogens.
4.1 Indicator Organisms
As previously discussed, testing for all forms of bacteria would be impractical due to the wide range of pathogenic bacteria found in the environment. To mitigate the time and cost of testing, laboratories examine bacteria whose presence indicates the possibility of food poisoning or the manifestation of other pathogenic bacteria (Forsythe, 2012).
These bacteria, also known as “indicator organisms”, are significant when assessing the microbiological safety and quality of foods (Forsythe, 2012). Indicator organisms are classified as either:
- ‘index’ organisms whose presence suggest the possibility of other pathogens being present; or
- ‘indicator organisms’ whose presence dictates the hygienic quality of the food (Forsythe, 2012).
In water testing, E. coli is used as the common indicator organism to determine the presence of enteric pathogens (Forsythe, 2012). Enumeration techniques are used alongside this method to count the number of organism’s present.
4.2 Classical Enumeration Methods
The type of enumeration method used will depend on the food or surface that is being tested and the specific organism that is being targeted. The following are enumeration methods that are commonly practiced in the food industry.
4.2.1 Plate Counts
In this method, agar plates are used to identify the number of actively growing cells in a sample. Selective and differential media are generally used to isolate and identify specific organisms such as coliforms. Selective media allows certain organisms to grow by using selective agents such as antibiotics, dyes or chemicals, whereas differential media uses agents that change bacterial growth patterns to differentiate groups of organisms (Otero Silva, Franceschini, Lavrador, & Candido, 2004).
In the food industry, non-fecal and fecal coliforms are grown on selective media such as Sorbitol MacConkey agar (SMAC) and Violet Red Bile agar (Forsythe, 2012). Food samples are employed on the agar by the ‘pour’ or ‘spread’ plate method as shown in Figure 3 (“A comparison between the pour plate method and the spread plate method | Scientific Diagram,” n.d.). In the ‘pour’ plate method, an empty plate is inoculated with the sample and melted nutrient agar is mixed with the sample. In the ‘spread’ plate method, the medium agar is pre-poured and allowed to solidify. Diluted samples are spread over the plate and incubated to allow growth (Forsythe, 2012).
Once incubated, colonies will grow on the medium, and the number of colony forming units (CFUs) are counted. The number of CFUs directly relate to the number of viable cells found in the sample. The success rate of this method is high, and these methods are cost-effective. The major drawback is the slow growth and long incubation times (Priyanka et al., 2016). 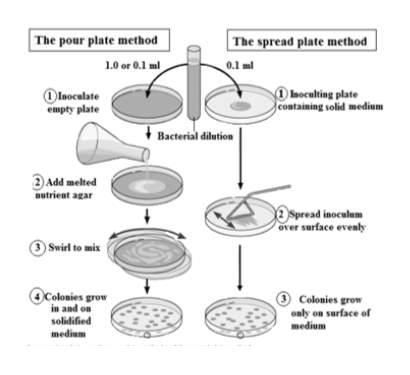
Figure 3: ‘pour’ plate and ‘spread’ plate method
4.2.2 Most Probable Number
The Most Probable Number (MPN) technique is used to estimate the number of viable microorganisms in a sample by using liquid broth and ten-fold sample dilutions. In this technique, five test tubes of broth are inoculated with 10 mL, 1 mL, and 0.1 mL of sample. The MPM can be determined by referencing the number of broths that show acid and gas production after 24-48 hours of incubation against statistical tables.
The second stage is to confirm the presence of coliforms by subculturing all the broths showing acid production or suspected colonies from agar into Brilliant Green Bile Broth (BGBB) (Forsythe, 2012). These tubes are incubated at 37 ⁰C for 48 hours (Forsythe, 2012). The presence of gas in this medium confirms the presence of coliforms. To confirm the presence of E. coli, BGBB can be used, but with an incubation temperature of 44 ⁰C for a 24 hour period (Forsythe, 2012). At the same time, a tube of peptone water is inoculated and incubated at 44 ⁰C for 24 hours (Forsythe, 2012). E. coli is one of the only organisms that can produce gas from lactose and indole from peptone at 44 ⁰C, so positive results confirm the presence of E. coli (Forsythe, 2012). This process is summarized in Figure 4. 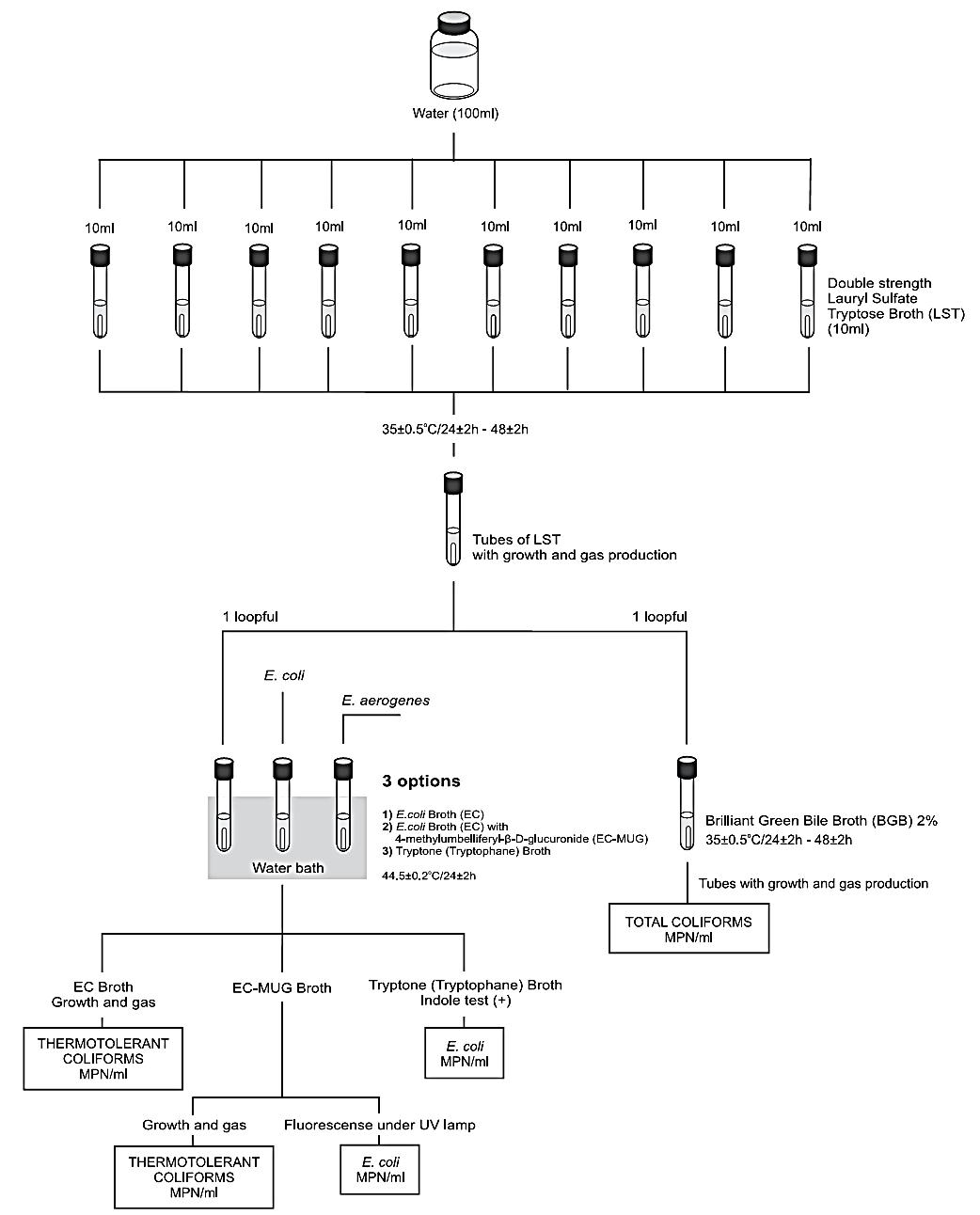
Figure 4: Flowsheet of MPN test for E. coli.
4.3 Dye-Reduction Test
The dye-reduction test is specific to the enumeration of bacteria found in milk. The two dye-reduction tests used in industry are the Methylene Blue Reduction test and Resazurin Test. The tests are based off the speed at which the dye disappears when added to milk. The colour disappears due to the removal of oxygen and the formation of reducing substances during bacterial metabolism. It is assumed that the greater the number of bacteria in milk, the quicker the oxygen is consumed and the faster the dye will disappear (“Dye Reduction Tests: Methylene Blue and Resazurin | Food Science,” n.d.).
In the methylene blue reduction test, 10 ml of milk is added to 1 ml of methylene blue thiocyanate solution in a test tube. The temperature of the tubes is increased to 36 ⁰C and the tubes are inverted every hour. If the milk remains colorized after 8 hours, the bacteria count is very low. On the other hand, if the milk decolorized before 2 hours, the bacteria count is very high (“Dye Reduction Tests: Methylene Blue and Resazurin | Food Science,” n.d.)
4.4 Immunological Methods
Immunoassays can detect specific proteins or other substances through their properties as antigens or antibodies. Enzyme-linked immunosorbent assay (ELISA) is the most common immunoassay used for pathogen detection (Priyanka et al., 2016). An advantage of ELISA is that the substrates will bind to their respective conjugates. As a result, each substrate will emit a different wavelength and the colours produced can be read on an ELISA reader (Priyanka et al., 2016).
However, the binding of the substrate and conjugate is very precise, and contamination can induce false positive results (Priyanka et al., 2016). Different substrates can be used to detect specific pathogens. For instance, to detect E. coli, pNPP was used as the substrate (Priyanka et al., 2016). ELISA has also been modified to account for other antigens and detection of other pathogens (Priyanka et al., 2016). For instance, the double antibody sandwich- ELISA (DAS -ELISA) is used to detect chicken flocks infected with S. enteritidis (Priyanka et al., 2016).
4.5 Polymerase Chain Reaction-Based Methods
In the polymerase chain reaction (PCR), specific genes of pathogens can be amplified and studied (Priyanka et al., 2016). The basic idea behind PCR is that genes are denatured by heat, annealed, and extended when the temperature is raised. PCR is faster than culture-based methods and immunoassays, which is advantageous for the identification of pathogens in food safety laboratories (Priyanka et al., 2016).
Although PCR is promising for the detection of genes in pathogens, the major drawback of this method is its high cost. (Priyanka et al., 2016). Additionally, PCR methods can have inconsistent results due to contamination, competing DNA in non-target cells, and cell lysis (Priyanka et al., 2016). The PCR technique has been modified specifically for the detection of foodborne pathogens (Priyanka et al., 2016).
In one of the methods, primers were developed for specific gene fimA of Salmonella and gen afa of E. coli to detect Salmonella and pathogenic E. coli (Priyanka et al., 2016). Real-time PCR has also been combined with the dye SYBR to detect strands of E. coli.
5.0 Microbial Quality of Food
The number of allowable pathogens found in food varies based on the type of food being analyzed. Based on government standards, a type of microorganism, group of organisms, or toxin produced by a microorganism, is specified to either not be present at all or will be allowed up until a certain concentration. For instance, Table 1 outlines the microbiological meat standards in Europe (Todd, 2004). Table 1: European Standards for pathogenic microorganisms in meat.
| Meat products | Samples taken | Meat preparations | Minimum | Maximum |
| Salmonella | 5 | 0 | - | - |
| E. coli | 5 | 2 | 50 per g | 500 per g |
| S. aureus | 5 | 2 | 100 per g | 1000 per g |
There exist standards for each food and pathogen that must be met to guarantee safe and quality food for consumption.
6.0 Environmental Monitoring Standards
Similar to food, the environment (e.g. a process control line, or a food processing factory) must abide by government standards to ensure optimal food-safety and decrease the risk of foodborne illness. These standards vary based on the location of the establishment, the food produced, and the equipment used.
6.1 Hazard Analysis & Critical Control Points
Hazard Analysis & Critical Control Points, also known as HACCP, is a program that controls potential biological, physical, and chemical hazards in food. This program ensures that food products are safe from food-safety hazards by monitoring and enforcing quality controls in food safety (da Cruz, Cenci, & Maia, 2006). The principles behind HACCP plans are conducted as follows:
- conduct a hazard analysis;
- determine the critical control points;
- establish critical limits;
- establish monitoring procedures;
- establish corrective actions;
- establish verification procedures; and
- establish documentation procedures (da Cruz et al., 2006).
6.2 Good Manufacturing Practices
Good Manufacturing Practices (GMP) provides the foundation for HACCP in an overall food safety programme (da Cruz et al., 2006). The topics relate to the environmental processing line, which includes cleaning, personnel and training, pest control, etc. (da Cruz et al., 2006).
6.3 Sanitation Standard Operating Procedures
Sanitation Standard Operating Procedures (SSOPs) are written procedures that ensure sanitary conditions of a food plant (da Cruz et al., 2006). They include:
- a description of operational procedures for sanitization;
- specification of the frequency of the procedures administered;
- identification of the individuals responsible for the implementation and monitoring of the SSOP; and
- the signature and date of the individual (da Cruz et al., 2006). SSOPs should be implemented along with the GMP programme (da Cruz et al., 2006).
7.0 Conclusion and Future Prospects
Microorganisms are found in abundance in the environment, especially in food and water. Some microorganisms can be beneficial to humans (in this report, the focus was placed on the process of fermentation) and others pose health risks to humans. In the food industry it is important to monitor food processes to optimize processes, reduce expenses, decrease the risk of food-borne illness, and maintain food standards outlined by HACCP. It is important to note that detection and monitoring technologies are constantly being improved and developed. This area of research and experimentation will continue to evolve to make technologies the most rapid, sensitive, specific, and cost-effective as physically possible.
8.0 References
A comparison between the pour plate method and the spread plate method | Download Scientific Diagram. (n.d.). Retrieved November 18, 2018, from https://www.researchgate.net/figure/A-comparison-between-the-pour-plate-method-and-the-spread-plate-method_fig6_257380059 Adams, M. R., Moss, M. O., & McClure, P. (2016). Food Microbiology. Royal Society of Chemistry. Alberts, B. (2017). Molecular Biology of the Cell. Garland Science. Batt, C. A. (2014). Encyclopedia of Food Microbiology. Academic Press. da Cruz, A. G., Cenci, S. A., & Maia, M. C. A. (2006). Quality assurance requirements in produce processing. Trends in Food Science & Technology, 17(8), 406–411. https://doi.org/10.1016/j.tifs.2006.03.003 Dye Reduction Tests: Methylene Blue and Resazurin | Food Science. (n.d.). Retrieved November 17, 2018, from https://www.uoguelph.ca/foodscience/book-page/dye-reduction-tests-methylene-blue-and-resazurin Filion, M. (2012). Quantitative Real-time PCR in Applied Microbiology. Horizon Scientific Press. Forsythe, S. (2012). Food Hygiene, Microbiology and HACCP. Springer Science & Business Media. Kurtzman, C., Fell, J. W., & Boekhout, T. (2011). The Yeasts: A Taxonomic Study. Elsevier. Lahtinen, S., Ouwehand, A. C., Salminen, S., & Wright, A. von. (2011). Lactic Acid Bacteria: Microbiological and Functional Aspects, Fourth Edition. CRC Press. McNeil, B., Archer, D., Giavasis, I., & Harvey, L. (2013). Microbial Production of Food Ingredients, Enzymes and Nutraceuticals. Elsevier. Otero Silva, J., Franceschini, S. A., Lavrador, M. A. S., & Candido, R. C. (2004). Performance of Selective and Differential Media in the Primary Isolation of Yeasts from Different Biological Samples. Mycopathologia, 157(1), 29–36. https://doi.org/10.1023/B:MYCO.0000012223.38967.7d Priyanka, B., Patil, R. K., & Dwarakanath, S. (2016). A review on detection methods used for foodborne pathogens. The Indian Journal of Medical Research, 144(3), 327–338. https://doi.org/10.4103/0971-5916.198677 Sanchez, P. C. (2008). Philippine Fermented Foods: Principles and Technology. UP Press. Todd, E. C. D. (2004). Microbiological safety standards and public health goals to reduce foodborne disease. Meat Science, 66(1), 33–43. https://doi.org/10.1016/S0309-1740(03)00023-8 Vojinović, V., Cabral, J. M. S., & Fonseca, L. P. (2006). Real-time bioprocess monitoring: Part I: In situ sensors. Sensors and Actuators B: Chemical, 114(2), 1083–1091. https://doi.org/10.1016/j.snb.2005.07.059
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Food and Nutrition"
Food and Nutrition studies deal with the food necessary for health and growth, the different components of food, and interpreting how nutrients and other food substances affect health and wellbeing.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




