Identification of Unknown Microorganisms
Info: 2687 words (11 pages) Dissertation
Published: 13th Oct 2021
Tagged: BiosciencesGenomics
Tradition Biochemical Techniques Contrast With 16s rRNA Gene Sequencing Method
Abstract
Identification of unknown microorganisms is necessary to understand how ecosystem function, remain diverse and result in reprocessing necessary elements for the ecology to remain in balance. This study focused on the use of traditional biochemical test and 16S rRNA molecular sequencing method to considerately evaluate the capacity that systems may facilitate the rapid and accurate identification of unknown bacteria. In gram-positive, one of the possibilities was offered by biochemical tests. In gram-negative bacteria, two of possibilities were given by biochemical tests, agreed with the results given by 16s RNA gene sequencing. 16S RNA sequencing method is more reliable and accurate for bacterial species that cannot be identified confidently by biochemical test alone and biochemical test can be used to validate 16s rRNA gene sequencing identification.
Introduction
Microbial species are abundant from upon above-surface to under-deep ground of the Earth and involve in multiples functioning to ecosystem services such as nitrogen cycle, plant growth and bioremediation[1],[2]. Soil is home of the most abundant species and experimental evidences showed biodiversity effects quantified ecosystem services and properties[3]. Also, extensive evidences suggest the loss of soil biodiversity subjectively affected ecosystem functionality and sustainability which society depend[4]. Identification of microorganism is one of foundation to approach their essential contribution to soil biodiversity, thus improving effects on ecosystem properties.
In this experiment, two unknow bacteria were predicted by examining traditional biochemical test and 16s rRNA gene sequencing method. Traditional biochemical test target to microbes' morphology and metabolic characteristic while 16s rRNA genes sequencing analysis can objectively assess phenotypic and genotypic characteristics of specific species. The species were predicted by integrating the accuracy and efficiency of both traditional biochemical test and 16s rRNA gene sequencing method and compared to the identity of the bacteria.
Material and Method
The more detail on standard experimental procedures is located on CBMS202/622 Practical Manual (2019)[5]
For incubation, all plates were incubated at 30oC overnight and stored in 4oC until next examination.

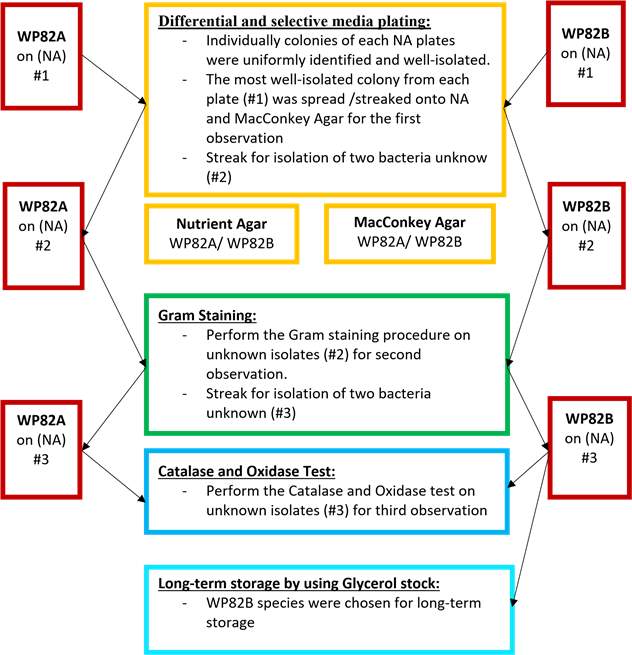

Results
*Observation of Bacillus thuringiensis refer to results on Practical 4 CBMS202/622 manual [5]
*Observation of Pseudomonas aeruginosa refer to [6]
Table 1. Biochemical Test Result Table 2. Theoretical Test Result
|
TEST |
WP82B |
WP82A |
Pseudomo nas aeruginos a |
Pseudomonas protegens |
Bacillus thuringiensis |
|
Nutrient Agar |
Circular, Convex, Entire, Cream colour |
Circular, Flat, Undulated, Cream colour. |
Bluish green colour |
Circular, Flat, Undulate, Cloudy cream. |
|
|
Growth on MacConkey Agar |
+ Gram (-) |
- Gram (+) |
+ Gram (-) |
+ Gram (-) |
- Gram (+) |
|
Gram Stain |
+ |
+ |
- |
- |
+ |
|
Catalase |
+ |
+ |
+ |
+ |
+ |
|
Oxidase |
+ |
+ |
+ |
+ |
+ |
|
Spore |
- |
- |
- |
- |
+ |
|
ONPG |
- |
- |
- |
- |
|
|
ADH |
+ |
+ |
+ |
+ |
|
|
LDC |
- |
- |
- |
- |
|
|
ODC |
+ |
- |
- |
- |
|
|
CIT |
+ |
+ |
+ |
+ |
|
|
H2S |
- |
- |
- |
- |
|
|
URE |
- |
variable |
variable |
- |
|
|
TDA |
- |
- |
- |
- |
|
|
IND |
- |
- |
- |
- |
|
|
VP |
- |
- |
- |
+ |
|
|
GEL |
+ |
+ |
+ |
+ |
|
|
GLU |
+ |
+ |
+ |
+ |
|
|
MAN |
- |
- |
- |
- |
|
|
INO |
- |
- |
- |
- |
|
|
SOR |
- |
- |
- |
- |
|
|
RHA |
- |
- |
- |
- |
|
|
SAC |
- |
- |
variable |
- |
|
|
MEL |
+ |
+ |
+ |
- |
|
|
AMY |
- |
- |
- |
+ |
|
|
ARA |
+ |
variable |
+ |
- |
Table 3. Top 3 BLAST result hits.
|
Organism |
% Query Cover |
E-Value |
%Ident. |
|
Pseudomonas protegens |
100 |
1 x e-49 |
93.79 |
|
Pseudomonas protegens |
100 |
1 x e-46 |
93.33 |
|
Pseudomonas aeruginosa |
46.8 |
0.30 |
79.05 |
Two gram-bacteria classification were observed on WP82P species at the beginning (Table 1). In addition, WP82B colonies appeared an alteration in diameter from small to large size after thirdstreaking plate, suggested us to further narrow down WP82B identification. According to biochemical test results, WP82B were suggested either Pseudomonas protegens, Pseudomonas aeruginosa or Bacillus thuringiensis (Table 1). WP82B and Bacillus thuringiensis were identified in the first round assuming gram-positive was reliable. However, morphological observation on Nutrient Agar and positive reaction on MacConkey Agar specifically indicated unrelatedness between WP82B and Bacillus thuringiensis (Table 1 and Table 2). Hence, WP82B were predicted to be gram-negative and thought either Pseudomonas protegens or Pseudomonas aeruginosa. Furthermore, biological media test reinforced that WP82B could not be Pseudomonas aeruginosa due to their physical bluish-green colonies onto MacConkey Agar(Table 2) [6]. Therefore, we put Pseudomonas protegens as our temporary prediction as stated by biochemical results.
To be more specific, as claimed by biomolecular sequencing test results, table 3 with three top BLAST result hits revealed their matching and confirmed our final prediction to be Pseudomonas protegens.
Discussion and Conclusion
The study was achieved with the aim of identifying microorganism that used many systemic processes to evaluate if such techniques might facilitate reliable prediction. Firstly, WP82B bacteria were cultured onto differential and selective medium to isolate and identify a particular organism. Members of WP82B grew, well-isolated and appeared as cream cloudy colonies, demonstrated WP82B organism can utilize nutrients source in the Nutrient Agar. In addition, on MacConkey Agar, members appeared reddy pink pigment demonstrated they ferment lactose to produce acid which results in pH decrease, suggested WP82B were gram-negative bacteria[7]. WP82B was stated positive result with the first Gram stain, however positive result onto
MacConkey Agar clearly evaluated a gram-negative result for WP82B (Table 1), suggested our first stain reaction was not properly investigated. This was thought due to the volume of the smear we used in Gram stain influenced the result of the decolourizing step[8]. To be more clear, another gram stain was reinvestigated, however, they still remained in positive gram reaction, stated their unreliability compare to biological media test. This was thought the age of the colonies may affect result due to our colonies selection was at the time of the alteration in size. To further narrow down species level, API20E was investigated and got 19 biochemical tests matched both Pseudomonas protegen and Pseudomonas aeruginosa. Bacillus thuringiensis was the only gram positive got 15 tests matched, however we rejected this prediction due to their uncommon morphology onto Nutrient Agar (Table 1). Therefore, gram negative dominantly could be predicted by applying API20E test and additional biochemical test were required to further investigate to species level[9].
The identification of WP82B was finally determined by using 16S rRNA sequencing methods. We did not obtain a good quality DNA sequence trace, they were quite noisy and week signal. However, the extracted region from the trace showed an extensive view of the bacterial culture, Pseudomonas protegens arrived on top 3 hits with 93.79% (Table 3) and we eliminated Pseudomonas aeruginosa due to their unfamiliar morphology.
Results above indicated 16S rRNA sequencing method is most reliable and accuracy for bacterial species that cannot be identified confidently by biochemical test alone. Biochemical techniques gave the impression of being variable, however, it can be used to validate 16s identification. Therefore, identification and classification of microorganism required the combination of efficiency both classical biochemical test and 16s rRNA genes sequencing, thus understand microorganism's biochemical ability and their genotypic variation. The factor is necessary to understand how ecosystem remains diverse and equilibrium.
In the future, time-efficiency and detection limits will be a key for the new expansion of investigating microorganism in order to interfere for improving dominant effects.
References
1. Venail PA, Vives MJ. Positive Effects of Bacterial Diversity on Ecosystem Functioning Driven by Complementarity Effects in a Bioremediation Context. PLoS One. 2013;8(9).
2. Burr TJ. Increased Potato Yields by Treatment of Seedpieces with Specific Strains of Pseudomonas fluorescens and P. putida. Phytopathology. 1978;68(9):1377.
3. American Journal of Science. Vol. 4, Science. 1896. p. 55–6.
4. Wagg C, Bender SF, Widmer F, Van Der Heijden MGA. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci U S A. 2014;111(14):5266–70.
5. Van Niel CB. Microbiology and molecular biology. Q Rev Biol. 1966;41(2):105–12.
6. Prasanna S, Dharanidevi S, Kumar Das N, Raj S. Prevalence, Phenotypic Characterization and Antibiotic Susceptibility of Non-Fermentative Gram Negative Bacilli Isolates at a Tertiary Care Centre. Int J Curr Microbiol Appl Sci. 2016;5(11):442–54.
7. Agar M. MacConkey Agar - Plate, Purpose, Ingredients and Principle | LaboratoryInfo.com. 2019; Available from: https://laboratoryinfo.com/macconkey-agar/
8. Cornell U. Gram stain protocols. Am Soc Microbiol [Internet]. 2005;(September 2005):14852. Available from: https://ahdc.vet.cornell.edu/docs/Gram_Stain_Protocol.pdf
9. Universidad Católica de Valparaíso., Chile. Comisión Nacional de Investigación Científica y Tecnológica. Electronic journal of biotechnology : EJB. 1997; Available from: https://www.researchgate.net/publication/320344807_Chable-Moreno_et_al_cultivo_de_embriones_de_maiz_CULTIVO_IN_VITRO_DE_EMBRIONES_INMADUROS_DE_CRUZAS_INTERPOBLACIONALES_DE_MAIZ_S2_CON_EL_EMPLEO_DE_BAP_Y_AIA_IN_VITRO_CULTIVATION_OF_IMMATURE_EMBRYOS_OF_
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Genomics"
Genomics is an interdisciplinary science that focuses on the study of all genes of an organism, their functions and how they interact with each other and their environment. The complete DNA, including all genes, of an organism is a genome.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




