Using CRISPR/CAS9 For Genomic Editing of the HIV-1 Virus
Info: 8137 words (33 pages) Dissertation
Published: 9th Dec 2019
Using CRISPR/CAS9 For Genomic Editing of the HIV-1 Virus
Hypothesis: Gene therapy when combined with HAART, will provide a way to effectively remove the HIV virus from the genome.
Abstract
Hypothesis
Gene therapy when combined with HAART, will provide a way to effectively remove the HIV virus from the genome.
Methods
The objective of this systemic review was to combine data from published literature that addresses the use of gene therapy to treat HIV. A search as conducted in Pubmed, Google Scholar, and NCBI using specific keywords related to the topic. Out of the hundreds of articles that were found, 13 articles were selected for this review. The data collected illustrated various experiments conducted using CRISPR/Cas9 on different cell types infected with the HIV-1 virus. The data collected focused on the effects of CRISPR/Cas9 on the HIV provirus as well as the ability of the system to eradicate latent HIV-1 provirus from the target cells.
Results
The CRISPR/Cas9 system is able to cause mutations in specific target sites of the HIV-1 provirus, creating cells that are resistant to HIV. It is also possible to excise the HIV genome from infected cells, leading to cell lineages that have effectively been cured of the virus.
Conclusions
CRISPR/Cas9 is a gene therapy technology that can be used in addition to HAART to treat and remove HIV from the genome.
Word count: 196
Keywords: CRISPR/Cas9, gene therapy for HIV, CCR5, latent HIV virus, stem cells, protein based site specific genomic editing
Introduction
The HIV epidemic has infected more than 70 million people worldwide since its discovery in 1983. Currently there are over 36 million people living with HIV/AIDS worldwide, with the highest prevalence in Sub-Saharan Africa (World Health Organization). The development of HAART has drastically improved the life expectancy of patients infected with HIV. HAART is the use of a multidrug regimen that acts on different viral targets. The goals of HAART are to achieve and maintain viral suppression, improve immune function and prevent opportunistic infections that often lead to death (Moore & Chaisson, 1999). The first antiretroviral drug used to treat HIV was a nucleoside reverse transcriptase inhibitor called zidovudine. However, by itself, it was unable to suppress the virus for long periods of time, resulting in patient deaths. The use of a combination of drugs was quickly incorporated into clinical trials and showed lots of promise for patients with HIV; there was a 60% to 80% decline in rates of AIDS, death, and hospitalization (Bai et al., 2013).
There are five classes of antiretroviral drugs used to treat HIV patients. These drugs are classified based on the lifecycle of the virus they inhibit. Fusion inhibitors prevent binding, fusion, or entry of HIV into the host cells by blocking various target structures on the host cell membranes. Nucleoside reverse transcriptase inhibitors are nucleoside analogs that prevent reverse transcription. Non-nucleoside reverse transcriptase inhibitors bind to allosteric sites on the enzyme to prevent reverse transcription. Protease inhibitors prevent the production of mature virions by stopping the virus from activating itself by inhibiting the protease necessary for cleavage of the polyprotein. Integrase inhibitors prevent the integration of the provirus into the human genome by inhibiting the viral integrase. There has been substantial progress made in anti retroviral therapies over the years. As of late 2006, the pill burden has been reduced to a single pill for once daily administration with the introduction of the pill efavirenz/tenofovir/emtricitabine. In addition, the development of CCR5 inhibitors and integrase inhibitors have garnered enthusiasm due to the impact they have on therapy but there are concerns about their efficacy and adverse effects, such as hepatotoxicity (Westby & van der Ryst, 2005).
Although HAART is very effective in treating patients with HIV, it does not fully restore health and there are problems associated with lifelong therapy. HIV is a disease that affects individuals on the genetic level. The biggest drawback to HAART is that it does not treat the underlying cause of this disease – the provirus embedded in the host cell’s genome. Adverse effects have been reported with the use of all antiretroviral drugs and are among the most common reasons cited for switching or discontinuing therapy and for medication non-adherence (O’Brien et al., 2003). Long-term treated patients are at a higher risk for a number of complications. For reasons that are not quite known, long-term treated HIV-infected patients have an expected life span that is substantially shorter than that of their HIV-uninfected peers (Antiretroviral Therapy Cohort Collaboration, 2008) and they have an increased risk for non-AIDS complications typically associated with aging, including cardiovascular disease, cancer, osteoporosis and other end-organ diseases (Deeks, S.G., 2011). The decrease in bone density of long-term treated HIV-infected patients is troubling because it has been projected that by 2015 more than half of the HIV/AIDS population in the USA will be over the age of 50 and the synergy between HIV and HAART-related bone loss with age-associated bone loss could lead to a significant health threat (Ofotokun & Weitzmann, 2010).
There is also evidence of adverse effects to HIV negative infants born to HIV positive mothers who are using anti-retroviral therapies. Fetal exposure to HAART causes impaired myocardial growth. These infants show reduced left ventricle mass, septal thickness and dimensions which are lower than normal. The overall loss of cardiac tissue with HAART exposure and an inability of the septum to grow could lead to left ventricle dysfunction (Lipshultz et al., 2011).
HAART is also very expensive; extending the length of life beyond 15 years after the initial diagnosis represents an accumulated cost of more than US $280,000.00 (Aracena – Genao et al., 2008). Another problem with the continual widespread use of HAART is the development of drug resistant strains of HIV. HIV is a retrovirus with no RNA polymerase proofreading activity, causing mutations to occur very rapidly, and as such, drug resistance will naturally occur. When HIV is not fully suppressed, drug resistance has a higher chance of occurring. This situation frequently occurs when there is a non-compliance with treatment (Colombrini et al., 2008).
It is clear that although the current therapies for HIV are effective for extending a patient’s life, they do not address the underlying causes of the disease. Once HAART is halted, individuals will rapidly rebound to their pre-HAART viral burden levels due to activation of proviruses from latent reservoirs (Hatano et al., 2000) and progression to AIDS occurs even after being treated for 5 years. Since the cause of the disease is at the genomic level, only genetic therapies can be used to effectively and reliably cure the disease. There are currently four techniques that can be used to cure HIV, one of which is the CRISPR/Cas9 system. These techniques can be used to recognize a specific DNA sequence and perform site specific gene editing.
The CRISPR/Cas9 system is a defense mechanism found in 40% of Bacteria and 90% of Archaea (Grissa et al., 2007). The system serves to protect against foreign genetic elements such as bacteriophages and plasmids that have previously infected the organism. It is the prokaryotic version of an adaptive immune system. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) are sequences in the prokaryotic DNA consisting of a succession of 24–47 bp repeated sequences separated by unique spacer sequences of similar length (Haft et al., 2005). The Type I-E CRISPR/Cas9 system, which is native to E. Coli, was the first CRISPR/Cas system that was identified. Most gene editing studies use a modified version of Type II CRISPR-Cas system, which is native to Streptococcus pyogenes (Jinek et al., 2012). CRISPR-associated (Cas) enzyme genes are often found in the CRISPR gene array. Currently there are 45 different types of Cas enzymes, of which only Cas 1 and Cas 2 are universally conserved (Haft et al., 2005). These enzymes mostly function as non-specific endonucleases and in the biogenesis of CRISPR-RNA.
The unique sequences found in the protospacer region between CRISPRs are foreign genetic material from phages and plasmids that have previously infected the organism. It seems that different CRISPR/Cas systems use different methods to acquire the spacer region (Horvath et al., 2012). Type I and Type II CRISPR-Cas systems rely on Cas1 and Cas2 to capture foreign genetic material while Type III does not (Yosef et al., 2012). Bioinformatics analysis of the spacer regions revealed that they were not randomly selected from the phage genome but were always adjacent to a short segment of DNA which became known as PAMs. PAMs, or Protospacer Adjacent Motifs, are 3 to 4 bp in length and play a role in the acquisition of foreign DNA and the maintenance of correct spacer length. They are also required for the specific targeting of Cas enzymes to foreign DNA. Cas enzymes won’t bind to the target DNA sequence if that sequence is not adjacent to a PAM (Shah et al., 2013). The bacteria use the PAMs as a means to distinguish self genome from non-self genome.
The CRISPR immune response involves two steps, synthesis of the CRISPR-RNA or crRNA and crRNA guided interference. The CRISPR array along with all the unique spacer sequences is transcribed as a single long mRNA. In type II CRISPR/Cas systems, this mRNA then base pairs with a trans-activation crRNA or TacrRNA to form a pre-crRNA/TacrRNA hybrid. This hybrid is then cleaved by RNaseIII and processed to form mature crRNA/TacrRNA or guide-RNA (Cong et al., 2013). This guide-RNA or gRNA contains one CRISPR and a unique spacer sequence complementary to a fragment of foreign genetic material. When a Cas enzyme associates with the gRNA hybrid, it is able to target the foreign genetic material that is complementary to the unique spacer sequence (Gasiunas et al., 2012).
There are several types of CRISPR/Cas systems that have been identified. Of these, the type II CRISPR-Cas system is the simplest CRISPR/Cas system, requiring only three components to function. The CRISPR/Cas9 system that is widely used in gene therapy is a modified version of the Type II CRISPR/Cas system. The TacrRNA and crRNA are ligated together to create a single piece of chimeric RNA, reducing the complexity to a two component system. In addition, the Cas9 nuclease has been optimized for human codons (Mali et al., 2013). This simplicity has made the type II CRISPR/Cas system more attractive for use in human gene therapy.
Methods
The articles reviewed in this paper were found with Pubmed, Google Scholar, and NCBI using specific keywords related to the topic. The keywords included: Genome editing; CRISPR/Cas9; HIV-1 integration; cure; experimental; gene therapy for HIV. For an article to selected for the systemic review, the following criteria had to be met:
- Publication date no older than 2013
- At least one experiment must have been done using the CRISPR/Cas9 system
- An animal model or cells created specifically as a substitute for human cells must have been used
- The HIV virus is the target of the experiment
- Discussion of advantages and disadvantages of using CRISPR/Cas9 for gene therapy
- Experimental data must clearly show the effects CRISPR/Cas9 had on DNA sequences
Certain exclusion criteria were also used in order to obtain articles with a focus solely on the CRISPR/Cas9 system. These criteria include:
- TALENS
- Zinc finger nucleases
- Meta-analysis studies
Articles which contained any of the exclusion criteria were excluded from this systemic review.
Data analysis was done using quantitative measures. Multiple pieces of literature were acquired related to using CRISPR/Cas9 for treating HIV using the specific criteria listed above. The data from the selected articles was then compiled and organized into an evidence table. The evidence table allows the reader to view the data collected in a clear and concise format. Within the table, the data is listed to show the type of study that was conducted, what the CRISPR/Cas9 system was tested on, and the results/outcomes of the therapy.
Results
The data collected from this systemic review demonstrates that the CRISPR/Cas9 system can be used to treat HIV as well as eliminate the HIV genes from the genome of infected cells. Zhu and colleagues (2015) demonstrated this effect by creating latent HIV cells that contained the full length viral DNA. 10 target sites for the Cas9 endonuclease were chosen, including 5 in the pol gene and 2 in the exon of the tat/rev gene. To demonstrate the effectiveness of the Cas9 endonuclease to cleave and mutate the HIV1 DNA at the target sites, the cells were treated with TNF-alpha to allow for viral expression, which was measured by the levels of GFP expression. Cells that were targeted by the Cas9 endonuclease showed a 5-fold decrease in GFP expression when compared to wild type cells that did not contain Cas9 (Figure 1A, 1B). When the levels of HIV-1 p24 antigen were measured using ELISA, viral production was decreased in DNA regions targeted by Cas9 compared to wild type cells (Figure 1C,1D). HIV-1 suppression was achieved in multiple HIV-1 DNA regions that were targeted by Cas9. Three different guide sequences (gRNAs) were cotransfected with Cas9 to recognize specific DNA sites within the genome of the HIV-1 virus. All 3 gRNA sequences included the target site for the tat/rev genes. When gene expression and viral production levels were analyzed in the gRNA sequences, there was a decrease in the number of GFP positive cells and a 24-fold decrease in viral production (Figure 1G, 1H). This finding demonstrates that the tat and rev genes of the HIV-1 virus can be specifically targeted by CRISPR/Cas9 to produce mutant HIV-1 viruses that have no viral activity.
Figure 2 shows that by using the CRISPR/Cas9 system to target the HIV1 LTR regions, it is possible to inhibit expression of the HIV1 provirus not only in the transcriptionally active provirus but also in the latent integrated provirus. Mutant HIV1 viruses were created, with 81.8% of the mutants containing a deletion in the LTR sequence while 9% of the mutants had insertions in the LTR sequence (Ebina et al., 2013).
CRISPR/Cas9 can target the latent HIV provirus in multiple types of human cells. Myeloid lineage cells are the primary cell types that harbor HIV-1 in the brain. Hu and colleagues (2014) showed that the HIV provirus can be targeted and disrupted in these cells. They designed a gRNA that targets the U3 region in the LTR. CHME5 microglial cells were transfected with a plasmid containing the gRNA and Cas9. Analysis of the cells showed high disruption efficiency of the genomically integrated HIV-1 provirus but there were some off target site cleavages. In Figure 3A, (right) it can be seen that the HIV-1 genome was successfully excised from chromosome Xp11.4. (Left) Using PCR, it is further seen that CRISPR/Cas9 was able to successfully remove the HIV-1 genome that was once integrated into the chromosomal DNA. They also demonstrated that Cas9/gRNA genome editing can be used to immunize cells against HIV-1 infection. In Figure 3B, it can be seen through PCR that cell clones containing Cas9 showed reduced expression of LTR regions where Cas9 excised the LTR sequence (Hu et al, 2014).
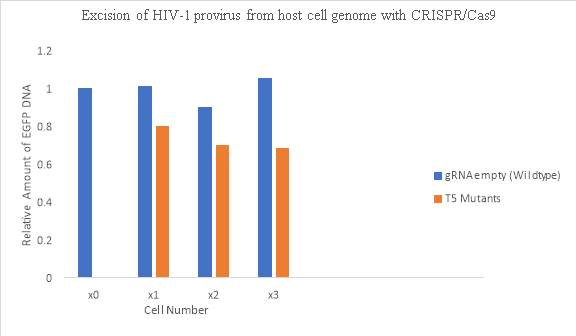
The HIV1 proviral DNA contains duplicate LTR regions on both ends of the integrated viral genome, allowing for CRISPR/Cas9 to cleave both LTR regions and remove the integrated viral genome from the host cell. In Figure 4, using quantitative PCR, CRISPR/Cas9 was again shown to reduce the LTR region GFP expression. The amount of DNA in the LTR region was decreased compared to the wild type cells not treated with CRISPR/Cas9. About 32% of the provirus was excised from the host cell genome after the CRISPR/Cas9 endonuclease was introduced into the mutant cells (Ebina et al, 2013).
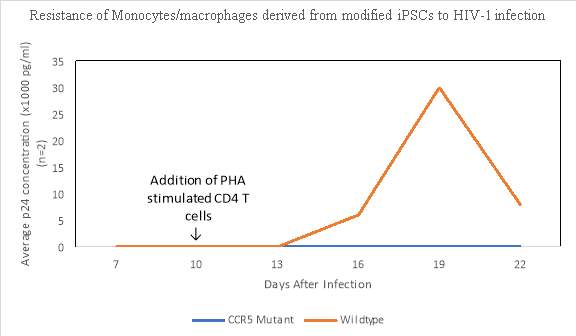
HIV resistant cells can be created by using lentiviral vectors expressing CRISPR/Cas9, allowing for the modification of HIV1 susceptible human CD4 T cells by disrupting the CCR5 gene making the T cells resistant to HIV 1 infection (Wang et al, 2014). Ye and colleagues (2014) also investigated whether a CCR5 mutant stem cell could be induced to produce monocytes and macrophages that were resistant to HIV-1 infection. After 24 days of culture, monocytes and macrophages (CD45+ CD14+ and CD14+ Cd11b+) were obtained from both parental wild type stem cells and the mutated cells infected with HIV-1. Phytohemagluttinin was used to stimulate the primary CD 4+ T cells and by day 16, HIV-1 viral replication could be detected in the monocytes and macrophages induced from the wild-type parental stem cells but no viral activity was detected in the cell lines derived from the mutated CCR5 stem cells (Figure 5). These results demonstrate that by using CRISPR/Cas9, pluripotent stem cells with a homozygous mutation for CCR5 can be created allowing for differentiation of monocytes and macrophages that are resistant to HIV-1.
Similarly, Liao and colleagues (2015) were able to engineer human-induced pluripotent stem cells that expressed HIV-targeted gRNA/Cas9. The modified stem cells differentiated into HIV reservoir cell types and showed resistance to HIV infection. This opens the possibility that a gene editing based vaccine strategy may be effective in eradicating integrated HIV-1 genome and newly packaged proviruses in cells.
Discussion
CRISPR/Cas9 is an endonuclease that recognizes a specific DNA site. When bound to the target site, cleavage of the DNA by the endonuclease creates double stranded DNA breaks, which can be repaired using nonhomologous end joining to produce insertions and deletions at the target DNA site, producing mutations within the target sequence (Zhu et al., 2015). By using CRISPR/Cas9, it is possible to create site specific mutations within the HIV-1 viral genome that results in reduced viral expression and viral production (Liao et al., 2015). This decrease in viral activity can be achieved in both transcriptionally active viral cells and latent viral cells integrated within the infected cell (Ebina et al., 2013). Mutations within the latent HIV cells prevents reactivation and infection of the virus (Hu et al., 2014). An important use of this ability of CRISPR/Cas9 to create mutations within the HIV-1 viral genome was demonstrated in multiple studies where CRISPR/Cas9 was used to create mutations in the CCR5 gene of the HIV-1 virus (Wang et al., 2014 & Li et al., 2015). Resultant clonal cells containing these mutations were found to be resistant to HIV-1.
The CRISPR/Cas9 system has been studied in many different cell types, including T cells and pluripotent stem cells infected with the HIV-1 virus. Based on all of the experiments that were analyzed for this systemic review, the data showed that by using CRISPR/Cas9 on HIV infected cells, it is possible to mutate the virus, leading to inhibition of viral replication and infectiveness. The cells with the mutant HIV virus can then produce daughter cells that are resistant to HIV. Thus, CRISPR/Cas9 can be used to treat and potentially eliminate the HIV virus from infected cells.
Patients that carry a homozygous deletion in the CCR5 gene have been reported to be resistant to HIV infection. CRISPR/Cas9 can be used to create a vaccine for HIV-1 that targets the CCR5 gene within T cells and macrophages. By using CRISPR/Cas9 to create site specific breaks in the CCR5 gene of T cells and macrophages, it is possible to create deletions within the gene, producing cells that are homozygous for the deletion in the CCR5 gene. Subsequent daughter cells will also contain the deletion in the CCR5 gene, resulting in daughter cells are effectively resistant to the HIV virus.
CRISPR/Cas9 can also be used to effectively excise the latent HIV-1 virus from the genome of the infected cells (Khalili et al., 2015). In a previous experiment, Mandal and colleagues (2014) were able to excise the CCR5 gene from CD 34+ hematopoietic stem cells. Clonal stem cell colonies all showed a deleted CCR5 gene making these cells immune to HIV-1. This finding suggests that it is possible to remove the HIV genome from infected cells, essentially curing the cells of the virus.
Transient suppression of genes can be used in combination with other gene therapies in the treatment of HIV. Qi and colleagues (2013) created a Cas9 mutant that lacks endonuclease activity. The mutant Cas9 can still bind to guide RNA to generate a DNA recognition complex that can specifically interfere with transcriptional elongation, RNA polymerase binding, or transcription factor binding. This system is called CRISPR interference (CRISPRi); it can be used to repress multiple target genes simultaneously, and its effects are reversible. It has been demonstrated that CXCR4 targeting CRISPRi causes significant suppression of CXCR4 in HeLa cells rendering them temporarily immune to CXCR4 tropic-HIV (Gilbert et al., 2013). By using CRISPR/Cas9, it may be possible to eliminate the HIV provirus from infected cells, while also producing uninfected stem cells that are immune to the virus, leading to a possible cure for HIV in the near future.
Limitations
The CRISPR/Cas9 system is a relatively new technology and there are some road bumps that need to be overcome. The major safety concerns that arise in gene therapy are off-target effects. What factors affect gRNA/Cas9 specificity is not fully understood. Some studies have shown that CRISPR/Cas9 can have substantial off-target effects. In vitro studies show that the Cas9 enzyme is lax in binding specifically at positions distal from the protospacer-adjacent motif region (Cong et al., 2013). The gRNA also only targets about 20 nucleotides; in large genomes, the lower specificity of the gRNA has the potential to create more off-target sites. Furthermore, the CRISPR/Cas system has been known to bind to DNA by non-Watson–Crick base pairing which results in unwanted mutations and cytotoxicity (Jiang et al., 2013). When Cradick and his team (2013) attempted to disrupt CCR5 in HEK-293 T cells using CRISPR/Cas9, they found that their CCR5-targeting CRISPR/Cas9 system induced off-target cleavage in CCR2, with mutation rates ranging from 5% to 20%.
Future Directions
The experiments included in this review studied the effects of CRISPR/Cas9 in vitro. The results obtained from these experiments have not been tested on human subjects. While the CRISPR/Cas9 system works in stem cells and T cells created specifically for the experiments conducted, the results may not be valid when these experiments are replicated in patients. Future studies need to demonstrate the effects of CRISP/Cas9 in clinical trials using human patients.
The major hurdle in gene therapy is target specificity and the potential for off target effects. Most gene editing approaches have some degree of off-target cleavage. However, gene therapy is still relatively new and it is constantly being improved upon and optimized for safe use in humans. The use of gene therapy has the potential to complement conventional antiretroviral therapies and enhance their effects. No clinical trials test the possibility of using a combination of gene therapy approaches in the treatment of HIV. Given the success achieved with combined drugs in HAART, it makes sense to devise treatment plans that use a combination of different gene therapy techniques along with HAART.
Conclusion
Given the current health and financial cost of the HIV epidemic, it is imperative to continue to pursue a variety of treatment strategies. Many protein and RNA based gene therapy options are available for use in the treatment of HIV infections. Although there are some short comings for the CRISPR/Cas9 system, it is a powerful technology for manipulating human genes. gRNA can easily and quickly be created at a very cheap price. Escape mutations by HIV won’t hamper this system as much as it would a system that uses a protein based domain for DNA recognition. CRISPR/Cas9 can also be used to target multiple strains of HIV. In addition, gRNA isn’t fused to Cas9 and Cas9 can function with all gRNAs, giving the system a great deal of flexibility. Given that it has only been a few years since CRISPR/Cas9 first started to be used for gene editing, it is very likely to be improved for both efficiency and to reduce off-target effects. From the data collected in this review, CRISPR/Cas9 looks promising for the treatment of HIV.
References
1. Aracena-Genao, B., Navarro, J. O., Lamadrid-Figueroa, H., Forsythe, S., & Trejo-Valdivia, B. (2008). Costs and benefits of HAART for patients with HIV in a public hospital in Mexico. Aids, 22, S141- S148. doi: 10.1097/01.aids.0000327635.74919.fd
2. Antiretroviral Therapy Cohort Collaboration. (2008). Life expectancy of individuals on combination antiretroviral therapy in high-income countries: a collaborative analysis of 14 cohort studies. The Lancet, 372(9635), 293-299. https://doi.org/10.1016/S0140-6736(08)61113-7
3. Bai, Y., Xue, H., Wang, K., Cai, L., Qiu, J., Bi, S., … & Liu, K. (2013). Covalent fusion inhibitors targeting HIV-1 gp41 deep pocket. Amino acids, 44(2), 701-713. doi:10.1007/s00726-012-1394-8
4. Colombrini, MRC, Coleta, D., Ferreira, M., & Lopes, MHBDM (2008). Risk factors for non- compliance with treatment with highly effective antiretroviral therapy. Revista da Escola de Enfermagem da USP , 42(3), 490-495.
http://dx.doi.org/10.1590/S0080-62342008000300011
5.Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., … Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (New York, N.Y.), 339(6121), 819–823. http://doi.org/10.1126/science.1231143
6. Cradick, T. J., Fine, E. J., Antico, C. J., & Bao, G. (2013). CRISPR/Cas9 systems targeting β- globin and CCR5 genes have substantial off-target activity. Nucleic Acids Research, 41(20), 9584–9592. http://doi.org/10.1093/nar/gkt714
7. Deeks, S. G. (2011). HIV Infection, Inflammation, Immunosenescence, and Aging. Annual Review of Medicine, 62, 141–155. http://doi.org/10.1146/annurev-med-042909-093756
8. Ebina, H., Misawa, N., Kanemura, Y., & Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9
system to disrupt latent HIV-1 provirus. Scientific reports, 3, 2510. doi:10.1038/srep02510
9. Gasiunas, G., Barrangou, R., Horvath, P., & Siksnys, V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 109(39), E2579–E2586.http://doi.org/10.1073/pnas.1208507109
10. Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., … Qi, L. S. (2013). CRISPR Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell, 154(2), 442–451. http://doi.org/10.1016/j.cell.2013.06.044
11. Grissa, I., Vergnaud, G., & Pourcel, C. (2007). The CRISPRdb database and tools to display
CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics, 8, 172. http://doi.org/10.1186/1471-2105-8-172
12. Haft, D. H., Selengut, J., Mongodin, E. F., & Nelson, K. E. (2005). A Guild of 45 CRISPR- Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Computational Biology, 1(6), e60. http://doi.org/10.1371/journal.pcbi.0010060
13. Hatano, H., Vogel, S., Yoder, C., Metcalf, J. A., Dewar, R., Davey Jr, R. T., & Polis, M. A. (2000). Pre-HAART HIV burden approximates post-HAART viral levels following interruption of therapy in patients with sustained viral suppression. Aids, 14(10), 1357- 1363. DOI: 10.1097/00002030-200007070-00008
14. Horvath, P., Romero, D. A., Coûté-Monvoisin, A.-C., Richards, M., Deveau, H., Moineau, S., … Barrangou, R. (2008). Diversity, Activity, and Evolution of CRISPR Loci in Streptococcus thermophilus . Journal of Bacteriology, 190(4), 1401–1412. http://doi.org/10.1128/JB.01415-07
15. Hou, P., Chen, S., Wang, S., Yu, X., Chen, Y., Jiang, M., … & Guo, D. (2015). Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Scientific reports, 5, 15577. DOI:10.1038/srep15577
16. Hu, W., Kaminski, R., Yang, F., Zhang, Y., Cosentino, L., Li, F., … & Mo, X. (2014). RNA- directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences, 111(31), 11461-11466. https://doi.org/10.1073/pnas.1405186111
17. Jiang, W., Bikard, D., Cox, D., Zhang, F., & Marraffini, L. A. (2013). CRISPR-assisted editing of bacterial genomes. Nature Biotechnology, 31(3), 233–239. http://doi.org/10.1038/nbt.2508
18. Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A
programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY), 337(6096), 816-821. DOI:10.1126/science.1225829
19. Khalili, K., Kaminski, R., Gordon, J., Cosentino, L., & Hu, W. (2015). Genome editing strategies: potential tools for eradicating HIV-1/AIDS. Journal of neurovirology, 21(3), 310-321. doi: 10.1007/s13365-014-0308-9
20. Li, Chang, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. Journal of General Virology 96.8 (2015): 2381-2393. DOI 10.1099/vir.0.000139
21. Liao, H. K., Gu, Y., Diaz, A., Marlett, J., Takahashi, Y., Li, M., … & Esteban, C. R. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nature communications, 6, 6413. doi:10.1038/ncomms7413
22. Lipshultz, S. E., Shearer, W. T., Thompson, B., Rich, K. C., Cheng, I., Orav, E. J., … & Starc, T. J. (2011). Cardiac effects of antiretroviral therapy in HIV-negative infants born to HIV-positive mothers: NHLBI CHAART-1 (National Heart, Lung, and Blood Institute Cardiovascular Status of HAART Therapy in HIV-Exposed Infants and Children cohort study). Journal of the American College of Cardiology, 57(1), 76-85. https://doi.org/10.1016/j.jacc.2010.08.620
23. Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., … Church, G. M. (2013). RNA- Guided Human Genome Engineering via Cas9. Science (New York, N.Y.), 339(6121), 823–826. http://doi.org/10.1126/science.1232033
24. Mandal, P. K., Ferreira, L. M., Collins, R., Meissner, T. B., Boutwell, C. L., Friesen, M., … & Musunuru, K. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell stem cell,15(5), 643-652. https://doi.org/10.1016/j.stem.2014.10.004
25. Moore, R. D., & Chaisson, R. E. (1999). Natural history of HIV infection in the era of combination antiretroviral therapy. Aids, 13(14), 1933-1942. doi:10.1097/00002030- 199910010-00017
26. O’brien, M. E., Clark, R. A., Besch, C. L., Myers, L., & Kissinger, P. (2003). Patterns and correlates of discontinuation of the initial HAART regimen in an urban outpatient cohort. JAIDS Journal of Acquired Immune Deficiency Syndromes, 34(4), 407-414. DOI: 10.1097/00126334-200312010-00008
27. Ofotokun, I., & Weitzmann, M. N. (2010). HIV-1 infection and antiretroviral therapies: risk factors for osteoporosis and bone fracture. Current Opinion in Endocrinology, Diabetes, and Obesity, 17(6), 523–529. http://doi.org/10.1097/MED.0b013e32833f48d6
28. Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., & Lim, W. A. (2013). Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell, 152(5), 1173–1183. http://doi.org/10.1016/jcell.2013.02.022
29. Shah, S. A., Erdmann, S., Mojica, F. J. M., & Garrett, R. A. (2013). Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biology, 10(5), 891–899. http://doi.org/10.4161/rna.23764
30. Wang, W., Ye, C., Liu, J., Zhang, D., Kimata, J. T., & Zhou, P. (2014). CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV –1 infection. PloS one, 9(12), e115987. https://doi.org/10.1371/journal.pone.0115987
31. Westby, M., & van der Ryst, E. (2005). CCR5 antagonists: host-targeted antivirals for the treatment of HIV infection. Antiviral chemistry and chemotherapy, 16(6), 339-354. https://doi.org/10.1177/095632020501600601
32. World Health Organization. http://www.who.int/gho/hiv/en/
33. Ye, L., Wang, J., Beyer, A. I., Teque, F., Cradick, T. J., Qi, Z., … & Levy, J. A. (2014).
Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proceedings of the National Academy of Sciences, 111(26), 9591-9596. https://doi.org/10.1073/pnas.1407473111
34. Yosef, I., Goren, M. G., & Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Research, 40(12), 5569– 5576. http://doi.org/10.1093/nar/gks216
35. Zhu, W., Lei, R., Le Duff, Y., Li, J., Guo, F., Wainberg, M. A., & Liang, C. (2015). The
CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology, 12(1), 1.
https://doi.org/10.1186/s12977-015-0150-z
Appendix
Evidence Table
| First Author | Date of Publication | Study Design | Level of Evidence | Study Population | Therapy or Exposure | Outcome/Results |
| Cradick, T. | August 11, 2013 | Experimental | 0 | Human Hb β gene and CCr5 gene | CRISPR/Cas9 | The CRISPR/Cas9 system does not have perfect specificity, resulting in off-target site mutations |
| Ebina, H. | August 26, 2013 | Experimental | 0 | HIV-integrated human cells | CRISP/Cas 9 Excision Therapy | HIV expression was reduced in both transcriptionally active and latent cells using the CRISPR/Cas 9 excision system |
| Hou, P. | October 20, 2015 | Experimental | 0 | Primary human CD4+ T cells, Jurkat cells, and Ghost-CXCR4 cells | CRISPR/Cas9 | Lentivirus mediated delivery of CRISPR/Cas9 into human CD4+ T cells can be used to target the CXCR4 gene of the HIV-1 virus, leading to mutations that confer resistance to the HIV-1 virus |
| Hu, W. | August 5, 2014 | Experimental | 0 | HIV infected myeloid cells in the brain | CRISP/Cas 9 system | CRISPR/Cas 9 system inactivated viral gene expression and replication in microglial, promonocytic, and T cells with latent HIV infection preventing reactivation and infection |
| Jiang, W. | September 01, 2013 | Experimental | 0 | E coli and S. pneumoniae | CRISPR/Cas9 | Using crRNA that is complementary to a target chromosomal sequence and a template DNA containing the desired mutation, CRISPR/Cas9 can induce mutations in targeted cells |
| Khalili, K. | June 21, 2015 | Experimental | 0 | HIV infected T cells and myeloid stem cells | CRISPR/Cas9 | Using CRISPR/Cas 9, HIV viral genome was eliminated from latent T cells and myeloid stem cells |
| Li, C. | August 5, 2015 | Experimental | 0 | HIV infected CD 4+ T cells | CRISPR/Cas 9 system | Using a recombinant adenovirus carrying CRISPR/Cas9, the CCR5 gene locus was cleaved to disrupt CCR5 expression resulting in HIV-1 resistance |
| Liao, H. | March 10, 2015 | Experimental | 0 | HIV infected pluripotent stem cells | CRISPR/Cas 9 system | CRISPR/Cas 9 was integrated into the genome of HIV infected stem cells. These cells showed decreased levels of viral replication over a period of 14 days |
| Mandal, P. | November 6, 2014 | Experimental | 0 | CD4+ T cells and CD 34+ stem cells | CRISPR/Cas 9 | CRISPR/CAS 9 was able to excise the B2M gene in CD 4+ T cells and the CCR5 gene from CD 34+ hematopoietic stem cells. The clonal stem cell colonies all showed a deleted CCR5 gene with a high rate of on-target mutations and a very low rate of off-target mutations |
| Qi, L.S. | February 28, 2013 | Experimental | 0 | E coli | CRISPR/Cas9 | A mutant CRISPR/Cas9 system can be created called CRISPR interference that silences transcription of specific gene without any off-target effects |
| Wang, W. | December 26, 2014 | Experimental | 0 | HIV infected CD 4+ T cells | CRISPR/Cas 9 lentivirus vectors | Cells with a CCR5 gene mutation were resistant to HIV infection and there were no detectable mutations in off-site regions |
| Ye, L. | July 1, 2014 | Experimental | 0 | Pluripotent stem cells | CRISPR/Cas 9 and TALENs | Pluripotent stem cells with a CCR5 mutation were created using the CRISPR/Cas 9 and TALEN system. These stem cells were found to be resistant to HIV infection. |
| Zhu, W. | February 27, 2015 | Experimental | 0 | HIV-1+ DNA | CRISPR/Cas9 | CRISPR/Cas 9 system was able to mutate the HIV viral genome at 10 target sites, preventing viral expression and virus production |
Figure 1. Zhu, W. et al (2015). Suppression of HIV-1 gene expression and viral production by gRNA/Cas9
Retrieved from https://doi.org/10.1186/s12977-015-0150-z
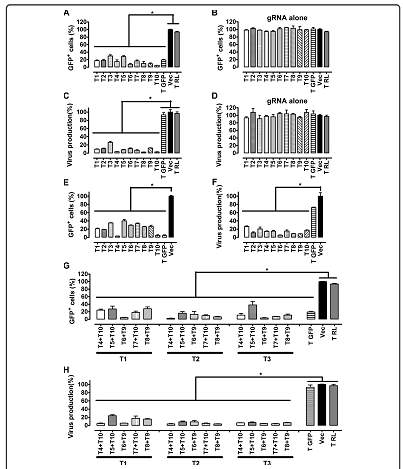
Suppression of HIV-1 gene expression and virus production by gRNA/Cas9. (A, B) Effects of gRNA/Cas9 on GFP expression. JLat10.6 cells
were transfected with gRNA and hCas9 plasmid DNA using Neon (Life Technologies). TNF-α (10 ng/ml) was added 16 hours after to stimulate HIV-1 gene expression from HIV-1 LTR promoter, which was monitored by flow cytometry. A gRNA targeting the GFP DNA (called T GFP (GTGAACCGCATCGAGCTGAA)) was included as a positive control. The gRNA targeting renilla luciferase DNA (called T RL (GTAGCGCGGTGTATTATACC)) was utilized as a negative control. Results obtained with the empty gRNA vector were arbitrarily set as 100%. The results shown are the average of three independent transfection experiments. Experiments were also performed with gRNA alone (without Cas9) and the results are shown in (B). (C, D) Effects of gRNA/Cas9 on virus production. Levels of viruses in the supernatants were determined by ELISA that measures HIV-1 p24 antigen. Results of three independent transfections are shown. (D)
represents the results of experiments performed with gRNA alone (without Cas9). (E, F) Pretreatment with TNF-α does not increase the inhibition of HIV-1 by gRNA/Cas9. JLat10.6 cells were treated with TNF-α (10 ng/ml) for 16 hours before nucleotransfection with gRNA and hCas9 plasmid DNA. GFP-positive cells were scored by flow cytometry and the results are shown in (E). Levels of viruses in the supernatants were determined by HIV-1 p24 ELISA and the results shown in (F). Results are the average of three independent transfections. (G, H) Suppression of HIV-1 by multiple gRNAs that target different HIV-1 DNA sites.
Three different gRNAs were co-transfected with hCas9 plasmid DNA. HIV-1 gene expression (shown in (G)) and virus production (shown in (H)) after TNF-α treatment were measured as described above. Results shown are the average of three independent transfection experiments. Asterisks indicate P < 0.05 (Mann–Whitney U test, SPSS 16.0).
Figure 2. Sequencing analysis of the CRISPR/Cas9-target site.
Data collected from: Ebina, H. et al. (2013). doi:10.1038/srep02510
A. Deletion
↓
WT: GGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTAACTAGGGAACCCA
WT: 9.1 % (2/22)
GGGTCTCTCTGGTTAGACCAGATCTGAG—TGGGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCCGA—— GGGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAG————————————-CTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCTGAG—–GGGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCTGA—CCTGGGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCTG————GGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCT—————-GAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCTGAG——————TCTCTGGCTAACTAGGGAACCCA
GGG———————————————————-AGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATC————CTGGGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGA—————————GCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCTGAG————–AGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGA———————————-TCTGGCTAACTAGGGAACCCA
Deletion: 81.8% (18/22)
B. Deletion and Insertion
WT: GGGTCTCTCTGGTTAGACCAGATCTGAG—————CCTGGGAGCTCTCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGATCTGAA—————————AGCTCGCTGGCTAACTAGGGAACCCA
GGGTCTCTCTGGTTAGACCAGAGATGTCAGCAGAGAGATGGGAGCTCTCTGGCTAACTAGGGAACCCA
Insertion: 9.0% (2/22)
Figure 3A. Hu, W. et al (2014). Cas9/LTR-gRNA efficiently eradicates latent HIV-1 virus from U1 monocytic cells. Retrieved from https://doi.org/10.1073/pnas.1405186111
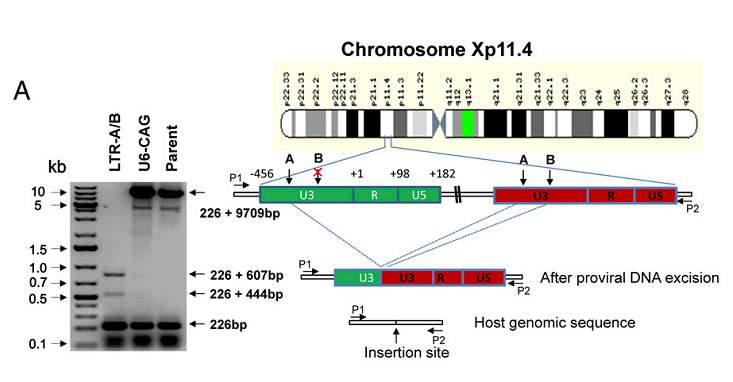
Cas9/LTR-gRNA efficiently eradicates latent HIV-1 virus from U1 monocytic cells. (A) (Right) Diagram showing excision of HIV-1 entire genome in chromosome Xp11.4. HIV-1 integration sites were identified using a Genome-Walker link PCR kit. (Left) Analysis of PCR amplicon lengths using a primer pair (P1/P2) targeting chromosome X integration site-flanking sequence reveals elimination of the entire HIV-1 genome (9,709 bp), leaving two fragments (833 and 670 bp)
Figure 3B. Stable expression of Cas9 plus LTR-A/B vaccinates TZM-bl cells against new HIV-1 virus infection. Data from Hu, W. et al (2014). https://doi.org/10.1073/pnas.1405186111
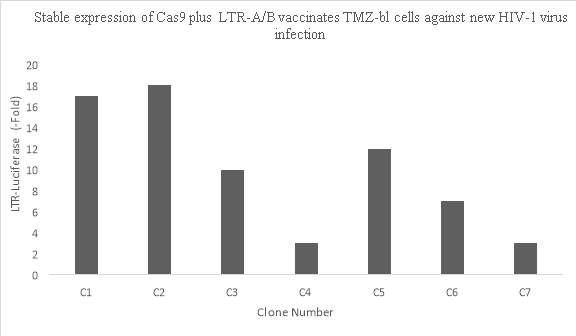
Figure 4. Excision of HIV-1 provirus from host cell genome with CRISPR/Cas9.
Data retrieved from Ebina, H. et al (2013). doi:10.1038/srep02510
j
Figure 5. Resistance of monocytes/macrophages derived from modified iPSCs to HIV-1 infection. Data retrieved from Ye, L. et al. (2014). https://doi.org/10.1073/pnas.1407473111
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Medicine"
The area of Medicine focuses on the healing of patients, including diagnosing and treating them, as well as the prevention of disease. Medicine is an essential science, looking to combat health issues and improve overall well-being.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




