Impacts of Inbreeding on the Two-toed and Three-toed Sloth
Info: 10290 words (41 pages) Dissertation
Published: 9th Dec 2019
Tagged: Biology
An integrated study into inbreeding and the impacts that arise when focusing on the two-toed and three-toed sloth.
ABSTRACT
This report investigates the effects of habitat fragmentation and the implications of inbreeding that arise when looking at B.variegatus and C.hoffmanni, two sloth species found in the neotropical forests of Costa Rica. 15 microsatellite markers have been used to analyse the genetic diversity of 150 individuals sampled across five separate populations across Costa Rica. Statistical tests revealed a low genetic diversity across all populations of sloth species sampled. Genetic diversity is critically low. However, there are little genetic divergences between each isolated population. It becomes unclear if habitat fragmentation is having a dramatic effect on the genetic diversity or if other factors are contributing. In this study, other factors have been observed to find that literature suggests polygamous mating systems are favoured in fragmented habitats. Further research would be necessary to investigate if there is a link between mating strategies and habitat fragmentation to determine whether this is causing low genetic diversity across individuals. Conservation strategies are discussed to be to understand how to regenerate an area once subjected to habitat loss and these strategies can be used to protect both B.variegatus and C.hoffmanni sloth species. If these issues continue to be ignored, it could lead to the extinction of an entire species and this research can be used and applied to the biodiversity of other species that are implicated by habitat loss and fragmentation.
INTRODUCTION
Sloths are members of the superorder Xenarthra along with armadillos and anteaters which have 31 extant species (Moraes-Barros et al, 2015). The Xenarthrans are believed to have evolved from other placental mammals approximately 100 million years ago, making them one of the oldest placental lineages (Moraes-Barros et al, 2015). At this time, they were a largely diverse group with approximately 218 genera (Moraes-Barros et al, 2015). This number has reduced significantly, and it remains unclear why. There have been previousmolecular phylogenetic studies (Moss et al, 2012) carried out and by combining these results with the data collected in this report, it can provide useful information on how processes such as habitat fragmentation have shaped the current genetic diversity in the superorder Xenartha but specifically the tree sloths (Folivora).
Choloepus hoffmanni, the two-toed sloth, and Bradypus variegatus, the three-toed sloth, are arboreal mammals which are located across the neotropical forests of Costa Rica (Moraes-Barros et al, 2006). The habitat for these species is crucial for survival as B.variegatus mainly lives on a diet of browsing leaves on trees whereas C.hoffmani feeds upon leaves and fruit (Montgomery & Sunquist, 1975; Taube et al, 2001). Both these species are listed as least concern in the IUCN red list of threatened species (IUCN, 2018). Sloths are one of the world’s most sedentary mammals with limited movement and have a reduced ability to maintain their body temperature (Cliffe et al, 2015). They rely solely on forest cover to provide food and shelter from other predators which leaves them highly susceptible to deforestation and habitat fragmentation (Moss et al, 2011). These sloths could offer an ideal model system for investigating dispersal rates and gene flow across Costa Rica as they are locally abundant in the habitat and can be handled easily(Moss et al, 2011).
Studies have been carried out on two other species of Bradypus in the Atlantic forest of Brazil, Bradypus torquatus and Bradypus pygmaeus which are both listed as threatened on the IUCN red list (IUCN, 2018). The Atlantic Forest has undergone 500 years of human impacts which have reduced the forest to 12% of the original coverage (Santos et al, 2015) and has left small, isolated areas for species to survive in. Both B.torquarus and B.pygmaeus species have been found to have a high population structuring and low levels of mtDNA diversity (Silva et al, 2017). As B.pygmaeus is isolated on a small island in Panama, studies have attributed this low genetic diversity to habitat isolation (Silva et al, 2017). Studies have also shown that B.vareigatus is sympatric with B.torquatus (Hayssen, 2010) and therefore being in close proximity to each other, similar findings for both species would be expected. Using previous studies on sloths across South America, it can be hypothesised that C.hoffmanni and B.variegatus would show similar characteristics of low genetic diversity when located in a fragmented habitat which is aimed to be investigated in this study.
The rapidly expanding human population has been driving deforestation to critical levels across the world. In Costa Rica these forests are heavily depended on by sloths and deforestation is destroying their habitats. Between 1940 to 1980 there was a rapid increasenumber of roads, farms, towns and cities now dominating the landscapes (Sader & Joyce, 1988) which can disrupt ecological processes and disrupt the species home range. Costa Rica has grown in tourism over the years and deforestation is occurring to accommodate this. By doing this, isolated segments are created which becomes detrimental to sloths who are limited by movement and drives habitat fragmentation. In this study, the aims are to investigate whether this rapid deforestation has had a devastating effect on the species inhabiting the forests of Costa Rica and if this has been the driving force of habitat fragmentation leading to reduced genetic diversity across sloth populations.
The Sloth Sanctuary in Costa Rica has rescued five baby sloths all of which show severe birth defects such as one finger and toe on each limb and albinism. When looking at populations, birth defects show a warning sign that inbreeding is possibly occurring. Birth defects can be a result of isolation on an island where inbreeding becomes the only method of reproduction available due to home ranges becoming isolated from other sloths. However, when comparing these sloths with birth defects from isolation, they are very similar to those found on the mainland suggesting that inbreeding may be occurring on mainland Costa Rica due to habitats becoming fragmented. This is of increased concern due to the potential impact that habitat fragmentation and inbreeding is having on the genetic health of wild sloth populations. This study aims to determine whether conservation strategies are needed immediately to protect the loss of sloth species in detrimental numbers and whole populations.
This investigation aims to use a non-invasive method for genetic sampling (Sharma et al, 2015) to determine whether habitat fragmentation is driving inbreeding across five sloth populations in Costa Rica. By using microsatellite markers, heterozygosity levels can be measured to determine the genetic diversity of each population and determine whether this is decreasing with the population size. Populations will also be analysed to investigate whether genetic distance correlates with geographical distance. This research also aims to look at the five rescued sloths showing severe birth defects and carry out microsatellite analysis to identify if these deformities are a result of inbreeding due to habitat fragmentation or are due to another possible cause.
The results from this study will provide an insight into the molecular diversity of two different sloth species located in Costa Rica and contribute to the knowledge of species diversity in the Costa Rican forests. The results may also provide conservation management strategies to prevent decline of an entire sloth population which may be due to habitat fragmentation and inbreeding.
MATERIAL AND METHODS
Herein 15 microsatellite markers for B.variegatus are obtained to use in genetic studies from Moss et al, 2011 and Moss et al, 2010. Hair samples from both species of sloths have been analysed to determine their diploid alleles which can be used to compare five separate populations against each other. Hair samples from the five sloth juveniles found with birth defects have also been examined to identify if homozygous alleles are present which could suggest inbreeding.
Data collection.
Hair samples were collected from 150 wild sloths in Costa Rica by Ph.D. student Rebecca Cliffe. These samples belong to B.variegatus species which were pooled into five main populations. 137 of these samples DNA were successfully extracted and amplified. 32 samples were collected from the population Limon, 28 samples were collected from Cahuita, 26 samples were collected from Penshurt, 27 samples were collected from Gualipes and 24 samples were collected from San Jose.
15 microsatellite markers were used and amplified in DNA. DNA was extracted from hairs of B.variegatus using a DNeasy extraction kit following the manufacturer’s protocol and amplified for 15 microsatellite markers from Moss et al, (2010) and Moss et al, (2011). Each marker is fluorescently labelled and has a unique number of base pairs and allele range which can be used for individual identification. In multiplex A the markers which were used were C121, A116, D3, B101, B109, B102, D2. In multiplex B the markers which were used were B119, D10, D107, D117, D1, D101, D9, B116.
For both multiplex A and B, 1.44ul of each forward and reverse primers were used and added to 96.96ul of sterile water. Primers were added to a mixture of 21.6ul of ddH2O and 72ul of master mix for amplification and 3ul of stock DNA was added to the reaction and the PCR products were run in an ABI1300 automatic sequencer at Aberystwyth University and analysed using Genemapper (Applied Biosystems, 2018).
Data analysis.
Each marker was rounded to its correct allele size depending on the number of nucleotides in their repeat sequence following Moss et al, 2010 and Moss et al, 2011. After the data for the alleles had been rounded, Genealex v6.503 (Peakall & Smouse, 2012) was used to carry out a range of statistical tests. Using Genealex (Peakall & Smouse, 2012), F statistics and polymorphism tests by population were carried out to provide values for the number of effective alleles, observed heterozygosity, expected heterozygosity and the fixation index for each population. An AMOVA test was also completed to provide global and pairwise FST values between populations to identify the level of genetic variance and if this is significant to 0.01. A Hardy Weinberg statistical test was also completed using the software Genealex (Peakall & Smouse, 2012) to discover if the data collected was significant to 0.01. A second software called Genepop on the web (Raymond & Rousset, 1995; Rousset, 2008) was also used to complete Hardy Weinberg analysis per population to the significance level of 0.01.
Using the pairwise FST values calculated between each population, an isolation by distance graph was also calculated using the Genealex software (Peakall & Smouse, 2012). This allowed the population FST pairwise values to be plotted against the geographical distance pairwise values to test the correlation between both genetic difference and geographical difference using a Mantel test.
The software Bottleneck (Cornuet & Luikart, 1997) was used to determine whether these five populations had been reduced to a small number and undergone a population bottleneck. The software ran three different statistical tests; the sign test, the standardised difference test and the Wilcoxon test. If these tests were significant, there is an increased chance of a bottleneck population to have occurred. The Bottleneck software (Cornuet & Luikart, 1997) also ran a test to detect if a mode shift had occurred between in any of the populations. A typical population would show an L shaped distribution in the software however when a shift away from this distribution occurs this indicates a bottleneck population to have occurred.
All the data collected was then arranged into figures and tables for further analysis and have been presented accordingly in the results section of this report to aid an answer for our hypothesis and discover if habitat fragmentation may be influencing the genetic diversity of our sampled sloths.
RESULTS
Figure 1 shows a pie chart created using the software Genealex (Peakall & Smouse, 2012) and carried out an AMOVA statistical test. The results show that there is a molecular variance within the individuals sampled at 53%. The results also show that there is a high molecular variance among the individuals sampled at 45% indicating there is genetic variance within and among the individual sloths sampled. However, when looking at the results between the five separate populations, there is only 2% molecular variance which is showing low genetic diversity between these populations.
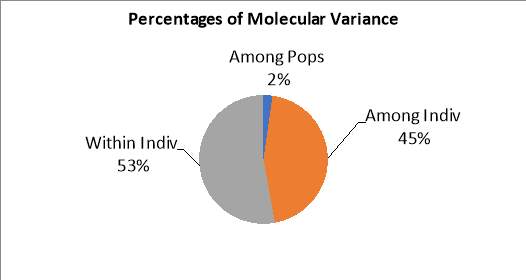
Figure 1: The percentage of molecular variance when looking at the individual and population level.
Table 1 shows the calculated heterozygosity, F statistics, and polymorphism by population for the data that has been collected. The NE column shows the mean number of effective alleles in each population which is rounded to 3 alleles across all five populations.
The next two columns show observed heterozygosity and expected heterozygosity. Overall the observed values of heterozygosity are lower than what was expected, suggesting that there are more homozygous individuals than first expected. However, heterozygosity values still range between 50-65% heterozygous individuals per population.
The final column shows the fixation index on a range from 0-1. The closer to 0, the higher the genetic material is being shared, and the closer to one means that no genetic material is being shared. These values are all low and suggest that genetic material is being shared between individuals.
Hardy Weinberg analysis is used to calculate P values for each population and determine whether these results are significant to 0.05. However, using the Bonferroni correction method, 0.05 is divided by the five populations to provide a significance level of 0.01. The results show that the populations of Limon, Penshurt and Gualipes are significant in Hardy Weinberg to a probability of 0.01.
| Pop | N | Ne | Ho | He | F | P | |
| Limon | Mean | 32 | 3.407 | 0.528 | 0.687 | 0.233 | 0.0000 |
| SE | 0.978 | 0.242 | 0.035 | 0.021 | 0.040 | ||
| Cahuita | Mean | 28 | 3.175 | 0.650 | 0.663 | 0.013 | 0.2278 |
| SE | 0.543 | 0.211 | 0.034 | 0.025 | 0.047 | ||
| Penshurt | Mean | 26 | 3.429 | 0.598 | 0.692 | 0.133 | 0.0004 |
| SE | 0.424 | 0.224 | 0.038 | 0.019 | 0.053 | ||
| Guapiles | Mean | 27 | 3.113 | 0.602 | 0.640 | 0.049 | 0.0189 |
| SE | 1.107 | 0.292 | 0.036 | 0.031 | 0.048 | ||
| San Jose | Mean | 24 | 3.154 | 0.511 | 0.659 | 0.217 | 0.0000 |
| SE | 0.502 | 0.234 | 0.030 | 0.025 | 0.045 |
Table 1: Sample size, number of effective alleles, observed heterozygosity, expected heterozygosity and fixation index for each sloth population.
By using an AMOVA statistical test with the software Genealex (Peakall & Smouse, 2012), an FST value for all the samples was calculated at 0.022 with a P value of 0.001 showing the data is significant. Values that were higher than 0.05 showed a genetic difference between individuals whereas if the value calculated was lower than 0.05 this indicates a low genetic difference between all samples. The value calculated was 0.022 suggesting a low genetic diversity between the individuals sampled.
Table 2 shows the pairwise FST values comparing each population genetic difference against each other. The results show that almost all the populations fall below 0.05 suggesting a low genetic difference overall. However, the populations Cahuita and Gualipes have a pairwise FST value of 0.07 indicating a medium genetic diversity between these two populations possibly due to fragmentation.
Table 2: The calculated pairwise FST values for each population where >0.05 shows a low genetic difference, 0.05-0.15 shows a medium difference and >0.25 indicates a high genetic difference.
| Limon | Cahuita | Penshurt | Gualipes | San Jose | |
| 0.000 | Limon | ||||
| 0.022 | 0.000 | Cahuita | |||
| 0.020 | 0.019 | 0.000 | Penshurt | ||
| 0.046 | 0.070 | 0.053 | 0.000 | Gualipes | |
| 0.025 | 0.046 | 0.041 | 0.031 | 0.000 | San Jose |
Figure 2 is showing genetic FST values plotted against geographical FST values to determine whether genetic difference increases as geographical distance increases. As seen in figure 2, a random correlation is showing no pattern between these two variables. Therefore, the genetic difference is not correlated to the geographical difference which would have been expected in a habitat that has not been affected by fragmentation.
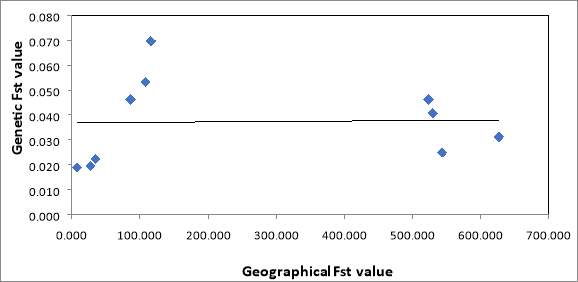
Figure 2: Isolation by distance graph to show genetic difference plotted against geographical difference which shows no correlation between the variables.
Table 3 shows the results of Bottleneck software that ran three tests; The sign test, the standardised differences test and the Wilcoxon tests. The results are shown in Table 4 for the Sign test and the Standardised difference test show that the populations Limon, Cahuita, Penshurst and Gualipes were significant to 0.01. When looking at the Wilcoxon test, all of the populations analysed were significant to 0.01 suggesting that they are likely to have undergone a population bottleneck.
Table 4 also shows the results when the software was used to detect if a mode shift had occurred. When looking at the results on a graph, a normal L-shaped distribution is expected however when a bottleneck population occurs, a shift from this graph is shown. The results show that only one population, Cahuita, shifted from the normal L shape expected and this suggests that this population may have undergone a bottleneck.
| Population | Sign test IAM | Standardised differences IAM | Wilcoxon test IAM |
| Limon | 0.00031 | 0.000086 | 0.000015 |
| Cahuita | 0.000232 | 0.000803 | 0.000015 |
| Penshurst | 0.000336 | 0.000607 | 0.000015 |
| Gualipes | 0.002637 | 0.001004 | 0.000107 |
| San Jose | 0.068852 | 0.010212 | 0.001068 |
Table 3:Results collected using Bottleneck software when using three different statistical tests; Sign test, standardised differences and Wilcoxon test.
| Population | Mode Shift | Bottleneck? |
| Limon | L Shaped | No |
| Cahuita | Shifted mode | Yes |
| Penshurst | L Shaped | No |
| Gualipes | L Shaped | No |
| San Jose | L Shaped | No |
Table 4: Results collected using Bottleneck software to determine if a mode shift has occurred which would indicate a bottleneck population.
DISCUSSION
Genetic diversity in B.variegatus.
Our results show that there is a low genetic diversity between the five populations sampled in this study. The AMOVA results show that there is a molecular variance of 53% within individuals and 45% among individuals (Figure 1) suggesting that there is genetic variation between each individual sampled. This shows that a high percentage of the sloths that were sampled are not sharing genetic material and inbreeding may not be occurring. However, the results from Figure 1 show that molecular variance between each population is only 2% showing that there is not a large genetic difference between the population of sloths in Costa Rica. Although these populations are fragmented from each other, the populations are shown to be genetically similar to each other which is uncommon in fragmented habitats. A study was carried out on the endangered Bradypus torquatus in the Atlantic forests of Brazil and it was found that the populations that were reproductively isolated and very divergent from each other. These divergent genetic clusters were specialised to different geographical locations across Brazil and it was found that this was due to allopatric fragmentation (Lara-Ruiz, Chiarello & Santos, 2008). The results found in this study show genetic divergence may be occurring between populations which therefore supports the hypothesis of this study.
When looking at the heterozygosity and F statistics shown in Table 1, the results show that the observed heterozygous individuals from each population deviated from the amount that was to be expected. This deviation from expected values suggests that the study found less heterozygous individuals in each population which may suggest inbreeding is occurring and genetic material is being shared. However, for each population, the results do not fall under 50% heterozygosity and therefore more than half of the individuals sampled in each population are heterozygous. This is still a high percentage of individuals which do not share the same genetic material and overall suggesting that there are more heterozygous individuals sampled than those that are homozygous. When looking at the fixation index calculated in Table 3, the closer the results are to 0 suggests that genetic material is being shared between the individuals within the population and each population is interbreeding freely. These values for each population are considerably low suggesting that genetic material is being shared between the individuals and interbreeding is occurring. When analysing the data using Hardy Weinberg, the data for the populations Limon, Penshurt and Gualipes were significant to 0.01 suggesting that the results found were not likely due to chance. The population Cahuita and Gualipes have extremely low values of 0.013 and 0.049 suggesting that these populations are sharing the most genetic material between each other and have the most genetic variance. This can also be linked to the high pairwise FST values between these two populations in Table 2 showing that there is a medium genetic difference between these populations.
Population structuring
When looking at the results collected by AMOVA for FST values, a similar pattern is discovered. The results show a calculated global FST value of 0.022 indicating low genetic diversity across all of the samples analysed. These results show that the individuals sampled overall are genetically similar suggesting that inbreeding may be occurring. When looking at the pairwise FST values shown in Table 2, these values are also significantly low and suggest a low genetic diversity between all the populations suggesting that genetic divergence is not occurring between these populations and going against the hypothesis. However, the only populations showing genetic variance are between Cahuita and Gualipes. These populations may be showing that isolation by fragmentation has occurred to a level that the populations are beginning to diverge from each other. These results suggest that four of the populations sampled are genetically similar to each other and divergence may not have occurred through habitat fragmentation and isolation. Habitat fragmentation started to occur across the forests of Costa Rica and Brazil 70 years ago and 4/5 of original cover had been lost (Zahawi, Duran & Kormann, 2015). Most of this habitat loss was in the first 30 years and for the past 50 years, regeneration has been occurring to grow secondary forest which is 30% of the original cover (Haddad et al, 2015). This could suggest that habitat fragmentation was an implication in the past and species have become isolated because of this. However, Costa Rica has been promoting forest regeneration since 1980 and this could explain why no genetic divergence is observed between the five populations sampled. However, habitat fragmentation may not be having a significant effect on biodiversity as once thought. Studies have been completed to investigate the Costa Rican dry forests which have been reduced to small patches which now only cover 0.1% of the original area (Janzen, 1989). A recent study investigated habitat patches in Costa Rica and found that the small area did not explain the high number of species found living there (Barrantes et al, 2016). The study suggested other factors such as environmental heterogeneity and complex topography may be influencing the number of species found instead. Alongside this study, it was also found that complex topography often had a higher species richness found than those patches with homogeneous topographies (Primack, 1998) and that species distribution may be more related to differences in vegetation rather than geographical isolation (Barrantes et al, 2016). Therefore, the habitats studied in this report may be fragmented but this does not necessarily mean that species richness and diversity is affected by this suggesting other factors need to be considered to provide an answer to the hypothesis in question.
Isolation by distance.
In Figure 2 the results show that there is no correlation between the genetic difference of each population and how geographically different each population is. In this study, it was hypothesised that the furthest populations from each other in geographical distance would also be the most genetically different. However, the results show that there is no relationship between these variables and each population has a similar genetic difference even though they are not geographically similar. The results show that four out of the five populations have a FST pairwise value of 0.02-0.05 with only one population having an FST value of 0.07. Therefore, from these results collected it can be suggested that four of the populations have a very low genetic difference which does not relate to the geographical difference. When looking at previous studies, habitat fragmentation is consistently associated with a decline in biodiversity (Clare & Horn, 2001). The area size of these small populations is one of the main factors that constricts populations as it is related to the amount of food available and competition for these limited food resources is high (Arroyo-Rodriguez & Dias, 2009). This becomes difficult for a population to survive, especially a mammal such as the sloth who has a solitary life, diet of specific plants and a low dispersion capacity (Lara- Ruiz & Chiarello, 2005). These changes are significant and change the population dynamics from the original population before fragmentation leading to a loss in genetic diversity and increased amount of inbreeding occurring (Lara- Ruiz et al, 2008). This study can support the results found in this study as genetic difference across all the populations is very low with an FST value of 0.02 which could suggest inbreeding is occurring due to habitat fragmentation. However, the genetic difference is low across all of the populations and there is still 50-60% heterozygosity in all of these populations going against our hypothesis that inbreeding is not occurring between populations and other factors may be causing deformities in the baby sloth rescued. Further studies show that habitat fragmentation will reduce reproductive success and the ability to adapt to the geographical environmental changes (Frankham, 2003; Lynch & Lande, 1998; Tabarelli et al, 2005). Not all the fragmented populations will have the same environmental conditions and therefore if populations were isolated, genetic differences between these populations would be expected. Schetino, Coimbra & Santos, 2017 studied fragmented habitats of the B.torquatos in the Atlantic forests of Brazil and found that there were genetic differences between the populations studied. They also found that the original populations were stable until anthropogenic deforestation and fragmentation had occurred approximately 50 years ago (Chiarello & Moraes- Barros, 2014). Therefore, in the results of the study, it was hypothesised to observe genetic variation between each fragmented habitat and this was only observed between the populations of Cahuita and Gualipes suggesting that there are possible other factors that may be causing this low genetic diversity shown. To support these results found, studies have been carried out to show that a surprisingly high level of biodiversity has been present for a variety of species undergoing habitat fragmentation in a reduced area (Mendenhall et al, 2014). Avifauna has been found to still have a high number of species present in the fragmented habitat of the Atlantic forests of Brazil (Daily, Ehnlich & Sanchez- Azofeifa, 2001). Other studies show comparable results for the vertebrate present in these fragmented habitats, especially bat species (Estrada & Coates-Estrada, 2001; Estrada, Cammarano & Coates- Estrada; Pulido-Santacruz & Renjifo, 2011). However, these studies specifically focus on animal species that can migrate from habitats by flying and therefore it cannot be applied to the sloth who is a sedentary mammal and is unable to migrate large geographical distances.
Evidence of bottlenecks
Further study was carried out to understand if any of the populations studied had undergone a bottleneck population due to areas being isolated into small fragmentations influencing a rapid decline in population numbers. The values shown in Table 3 show the results for three significant tests ran on each population. Each population was significant in all test carried out apart from San Jose in the Sign test and the Standardised difference test. This suggests that four of these populations have undergone a population bottleneck. These results suggest that habitat fragmentation of these populations is isolating the individual sloths sampled to a point where the species number is declining. If habitat fragmentation were occurring on a serious scale, divergent genetic clusters would be expected to be observed which would be specific to each geographical region and this would be caused by allopatric fragmentation (Lara-Ruiz, Chiarello & Santos, 2008). Therefore, it may be suggested that these five populations have not undergone severe habitat fragmentation to the point where species are declining as the only populations to be observed as genetically divergent to each other is Cahuita and Gualipes. Other factors may be influencing some inbreeding between these species of sloths and this study shows that in the future conservation of these habitats needs to be of high priority to prevent the sloth species from undergoing a bottleneck population and their existence being threatened by genetic variation and habitat fragmentation. It is not just genetic diversity that is threatened in fragmented habitats. Other factors for population decline can include pesticides, hunting, fire and native dogs (Daily et al, 2003). A study found that the giant anteater, which is a relative of the sloths, is susceptible to fire and native dogs and therefore is unable to survive in fragmented habitats (Nowax, 1999). Sloth populations need to be undergoing conservation management immediately to prevent population decline to a critical level not only by reducing genetic diversity but also other factors such as hunting, wild dogs and forest fires. A final statistical test was carried out to determine further if any of the five populations sampled had undergone a bottleneck population.
The results in Table 4 are used to determine if a mode shift had occurred from a normal L shaped distribution. A mode shift indicates that a population may have undergone a population bottleneck. The mode shift results show that four of the populations sampled did not undergo a mode shift and therefore are unlikely to have undergone a bottleneck population. The results for the population of Cahuita show that a mode shift has occurred and that a bottleneck population may be present. This supports the results shown in table 2 as Cahuita and Gualipes have the highest pairwise value of 0.07 showing genetic diversity between these two populations suggesting that divergence has occurred by isolation due to habitat fragmentation. However, it can also be shown in Table 1 that the population of Cahuita has an observed heterozygosity value of 65% and a low fixation index of 0.013. This shows that there are a high number of heterozygous individuals in this population and that interbreeding is happening freely. This would not be expected to be shown in a bottleneck population where the number of species has been rapidly reduced so that species would have to inbreed and therefore our results may be an anomaly. In this case, it would expect to see very low heterozygosity expected values and a higher fixation index nearer to 1. In studies where habitats have been reduced 93% of their original size (Lara-Ruiz, Chiarello & Santos, 2008) these populations become reproductively isolated and therefore a high number of homozygous individuals would be observed in these habitats. However, this is not seen in the results collected in this study suggesting that habitat fragmentation is not effecting B.variegatus and C.hoffmanni species as heavily as once thought. Sloth species need high densities of shade and cover in their habitats to provide uninterrupted connectivity between trees (Chiarello et al, 2004). B.variegatus and C.hoffmanni have similar movements on the ground but differ when climbing larger trees and descending (Mendel, 1985). It is important to have as large gaps will stop sloth from moving between trees as they do not jump and are reluctant to use the ground due to elevated levels of predation and limited movement (Chiarello et al, 2004). This suggests that if habitat fragmentation has occurred in Costa Rica, that due to their sedentary lifestyle, sloths would be highly interbred, and this is not shown in the results of this study going against our hypothesis that habitat fragmentation is having a large effect on the population sizes and inbreeding of sloths. However, a study has observed that C.hoffmanni species can repeatedly venture into open pastures in search of preferred trees (Cassano, Kieruff & Chiarello, 2011). This derives from the sloth’s original anatomy of being strongly evolved for the arboreal lifestyle (Goffart,1971) and may suggest that sloths are becoming able to adapt to habitat fragmentation by moving across open pastures and while doing this they may be able to interbreed with other sloth individuals which would not first be possible. This could support the results found in this study that the habitats may be heavily fragmented, but this is not affecting the sharing of genetic material between the individual sloth species.
Mating systems of B.variegatus and C.hoffmanni.
To investigate this further, habitat fragmentation may not be the only factor that is contributing to low genetic diversity and inbreeding found in sloth individuals. The mating system of both B.variegatus and C.hoffmanni need to be critically analysed to determine if this is effecting the sharing of genetic material between individuals. Sloths are sedentary mammals with low metabolic rates and up to 14 hours of inactivity a day (Pauly & Peery, 2012). Due to their solitary nature and their small teste size, it is considered that competition should not be a predominant feature (Garcés – Restrepo et al, 2017). Living in an area with a small home range, it has been investigated that female promiscuity is the main mating strategy in sedentary mammals such as B.variegatus and C.hoffmanni (Pauly & Peery, 2012). This is believed to arise in mammals as a survival strategy as it can increase the genetic fitness of the offspring and reduce the risk of infanticide (Pauly & Peery, 2012; Emlen & Orign, 1977). Promiscuity is favoured in habitat fragmentations that are highly fragmented because there is also high competition between male species in these areas. High aggression has been observed between males in these areas (Greene, 1989) which is often observed in polygamous mating systems. It is even more likely to observe female promiscuity in sedentary species as males are unable to monopolise access to many mating opportunities (Pauly & Peery, 2012). However, these mating systems can be ambiguous. Mating systems could become flexible across time and change alongside resource availability (Pauly & Peery, 2012). A study carried out by Pauly & Peery found that B.variegatus species surprisingly possess a high polygamous mating system compared to the once thought promiscuity mating system. The study found that 16% of adult males gave rise to 85% of the juveniles in one habitat. This could support the results found in this study as inbreeding and low genetic diversity across all populations could be due to the overall mating systems of B.variegatus. It was also found that when habitats were fragmented, many females can persist in small patches of habitat and be monopolised by one single male (Pauly & Peery, 2012). This further supports the results found in this study as one male’s genes are monopolising many females. If this occurs across generations, the genetic material being shared between individuals will be very similar and therefore inbreeding may be occurring. It was also found that C.hoffmanni had a mixture of weak polygyny and female promiscuity (Peery & Pauly, 2012). Habitat fragmentation may have a small influence on the mating systems of sloths due to decreasing the number of female to male ratio in a small habitat area, increasing polygamy and inbreeding between individuals. However, it can be argued that sloth species are polygamous even if they are not in a fragmented habitat. The mating systems naturally lean towards male species monopolising many females and sharing the same genetic material between a population. This could support the results in this study and explain why genetic diversity between sloth species is low across all populations and all individuals and explain why there has been an increased number of juveniles showing severe birth defects being found due to the B.variegatus and C.hoffmanni having a polygamous mating system.
Conservation strategies.
To find a solution to the hypothesis in question, conservation strategies need to be in place across the forests of Costa Rica. Habitat fragmentation and loss of habitat are expected to be observed in a country relying heavily on an expanding population. Roads and expanding agricultural areas rely on the clearing of habitats to be able to support the expanding habitat and this is consistently linked to the decline of biodiversity of many species (Clement & Horn, 2001). Conservation strategies are crucial to the survival of B.variegatus and C.hoffmanni as they are arboreal mammals and species numbers would decline rapidly if the loss of habitat had occurred. In a recent study of Costa Rica, it was found that 80% of original forest cover had been lost (Zahawi, Duran & Kormann, 2015). This was primarily driven by the increasing demand for agricultural products across the country (Metzger et al, 2009; Gloor et al, 2012; Ceballos et al, 2015). However, in the present day, 30% of the forests are regenerated secondary forests. Looking at the results found in this study it can be observed that this is not enough as the sloth species of Costa Rica still have an overall low genetic diversity which could be due to loss of habitat. Daily et al, 2003 investigated the conservation of mammals in neotropical areas and found that 35% of the native species were living in a restricted habitat compared to 54% of the land being used for agricultural purposes. It has also been found that pesticides in these agricultural landscapes have been actively found in sloths which have caused acute and chronic effects (Branford et al, 2014). It was found that there is not a substitute for removing native forests and because of this, six species were extinct locally from the study area and many other species had reduced rapidly in size and density (Daily et al, 2003). This study stresses the importance of preventing habitat loss as it becomes detrimental for those species that rely so heavily on the native environment and it rapidly increases the loss of an entire species. A conservation method that could be easily implicated into the forests of Costa Rica is using cacoa agroforests as habitats for sloth species such as B.varigeatus and C.hoffmanni. A study found that there are many biologically rich cacoa agroforests that are merged with the natural landscapes of Brazil (Cassano, Kieruff & Chiarello, 2011). Supporting this further, Vaughan et al, 2007 carried out a study using both B.variegatus and C.hoffmanni in cacao agroforests embedded into agricultural landscape. It was found that C.hoffmanni were present in 101 tree species and B.variegatus were present in 71 tree species. This shows that these landscapes provide habitat for species which have lost their native habitat to fragmentation. These habitats are highly composed of native forest habitat which can be used as a habitat for the B.torquatus species providing a larger habitat for these mammals to exist and reproduce in which resembles closely their native habitats. A further study in Costa Rica has found that B.variegatus and C.hoffmanni are also able to use cocoa landscapes as a habitat (Goffart, 1971). These methods combined with a further study on the area could provide a successful conservation method to protecting the sloth species from declining and also provide the opportunity for genetic diversity between individuals to gradually increase. However, these methods would favour C.hoffmanni more as they have been found to live and survive in these areas due to greater adaption to habitat use (Mendoza et al, 2014). B.variegatus have difficulty surviving outside of their natural habitats as they do not adapt to substitute diets, they are susceptible to different temperatures and they have a much lower defence against common diseases (Taube et al, 2001). This method would require further action to prevent the extinction of a species by using techniques of active management alongside this cacao landscapes (Cassano, Kieruff & Chiarello, 2011). The main goals of achieving conservation rely on the maintenance of evolutionary processes alongside the preservation of functioning landscapes (Moritz, 2002; Altieri, 1999). By spending time regenerating and to study the ideal landscapes of sloth species further, landscapes can become protected from any damage by habitat loss and become the ideal environment for increasing biodiversity. It is also fundamental to maintain the fitness and adaptive capacity of the local populations to environmental changes (Lynch & Lande, 1998; Moritz, 1994). By adapting conservation strategies to fit the environmental changes and conditions of the landscape provides an ideal habitat for species to thrive in which will also remain to protect the biodiversity for many years. When looking at environmental changes, a study was completed by Moreia et al, 2014 to discover if the climate is related to species diversity. The study found that the models do not indicate that climatic relation with an absence of species. Other studies found that there were suitable climatic conditions even in areas where species were absent (Oliver & Santos, 1991; Gardner, 2007; Aguiur, 2004).
To provide a conclusion for this investigation, it can be shown that there is low genetic diversity between the sloth individuals sampled in this study. It is evident that there is a factor that is reducing this genetic diversity to low levels. Habitat fragmentation has been an ongoing issue across neotropical environments and has had critical implications of habitat loss which have led to a decline in biodiversity. In this investigation there were not clear genetic differences between all isolated populations, genetic diversity was low across the whole of Costa Rica. This data suggests that habitat fragmentation is affecting sloth populations as first hypothesised. However, other factors need to be investigated to determine the main cause of the low genetic diversity observed. The mating strategy of sloths may explain why genetic diversity is observed low across the neotropics of Costa Rica. Habitat fragmentation may change mating strategies of species to polygamous to continue genetic fitness. If one male is siring many of the offspring in one habitat, it can be expected that low genetic diversity levels will be observed. Further studies would need to be carried out on a larger scale to investigate the mating strategies of both B.variegatus and C.hoffmanni and determine whether habitat fragmentation is correlated to polygamous mating systems. Finally, conservation strategies discussed in this report would provide a foundation to begin protecting B.variegatus and C.hoffmanni. By regenerating forests and using cacao agricultural landscapes as secondary habitats for sloth populations while alongside protecting areas from habitat loss and deforestation, sloth numbers will be able to increase and there will be an increase of genetic diversity between individuals. Overall it is crucial to gain an understanding of what could be causing this decline in genetic diversity and, alongside further studies, this data can provide an insight to prevent the loss of both the B.variegatus and C.hoffmanni sloth species in the neotropical forests of Costa Rica.
ACKNOWLEDGEMENTS
This research was carried out at Swansea University College of Science as an undergraduate student under the supervision of Professor Sonia Consuegra del Olmo. I highly acknowledge Ph.D. student Rebecca Cliffe for providing the DNA samples for analysis in this study and Ph.D. students Ben Whittaker and Chloe Robinson for their help and guidance of the software used for microsatellite analysis.
REFERENCES
Aguiar J (2004) Species Summaries and Species Discussions. Edentata, 6, 3-26.
Altieri M (1999) The ecological role of biodiversity in agroecosystems. Agriculture, Ecosystems & Environment, 74, 19-31.
Arroyo-Rodriguez V, Dias P (2010) Effects of habitat fragmentation and disturbance on howler monkeys: a review. American Journal of Primatology, 72, 1-16.
Barrantes G, Ocampo D, Ramírez-Fernández J, Fuchs E (2016) Effect of fragmentation on the Costa Rican dry forest avifauna. PeerJ, 4, e2422.
Branford M, de la Cruz E, Solano K, Ramirez O (2014) Pesticide exposure on sloths (Bradypus variegatus and Choloepus hoffmanni) in an agricultural landscape of Northeastern Costa Rica. Journal of Environmental Biology, 35, 29-34.
Cassano C, Kierulff M, Chiarello A (2011) The cacao agroforests of the Brazilian Atlantic forest as habitat for the endangered maned sloth Bradypus torquatus. Mammalian Biology – Zeitschrift für Säugetierkunde, 76, 243-250.
Ceballos G, Ehrlich P, Barnosky A, Garcia A, Pringle R, Palmer T (2015) Accelerated modern human-induced species losses: Entering the sixth mass extinction. Science Advances, 1, e1400253.
Chiarello A, Chivers D, Bassi C, Maciel M, Moreira L, Bazzalo M (2004) A translocation experiment for the conservation of maned sloths, Bradypus torquatus (Xenarthra, Bradypodidae). Biological Conservation, 118, 421-430.
Chiarello AG, Moraes-Barros N (2014) Bradypus torquatus. The IUCN Red List of Threatened Species Version 20142. IUCN Red List.
Clement R, Horn S (2001) Pre-Columbian land-use history in Costa Rica: a 3000-year record of forest clearance, agriculture and fires from Laguna Zoncho. The Holocene, 11, 419-426.
Cliffe R, Haupt R, Avey-Arroyo J, Wilson R (2015) Sloths like it hot: ambient temperature modulates food intake in the brown-throated sloth (Bradypus variegatus). PeerJ, 3, e875.
Cornuet J.M. and Luikart G., 1997 Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144, 2001-2014.
Daily G, Ceballos G, Pacheco J, Suzan G, Sanchez-Azofeifa A (2003) Countryside Biogeography of Neotropical Mammals: Conservation Opportunities in Agricultural Landscapes of Costa Rica. Conservation Biology, 17, 1814-1826.
Daily G, Ehrlich P, Sánchez-Azofeifa G (2001) Countryside Biogeography: Use of Human-Dominated Habitats by the Avifauna of Southern Costa Rica. Ecological Applications, 11, 1-13.
Emlen S, Oring L (1977) Ecology, sexual selection, and the evolution of mating systems. Science, 197, 215-223.
Estrada A, Coates-Estrada R (2001) Bat species richness in live fences and in corridors of residual rain forest vegetation at Los Tuxtlas, Mexico. Ecography, 24, 94-102.
Estrada A, Cammarano P, Coates-Estrada R (2000) Biodiversity and Conservation, 9, 1399-1416.
Garcés-Restrepo M, Peery M, Reid B, Pauli J (2017) Individual reproductive strategies shape the mating system of tree sloths. Journal of Mammalogy, 98, 1417-1425.
Gardner AL (2007) Mammals of South America: Volume I. Marsupials, Xenarthrans, Shrews, and Bats. University of Chicago Press, Chicago.
Genemapper V 4.0 (2018) Applied Biosystems, California.
Gloor M, Gatti L, Brienen R et al. (2012) The carbon balance of South America: a review of the status, decadal trends and main determinants. Biogeosciences, 9, 5407-5430.
Greene H (1989) Agonistic Behavior by Three-toed Sloths, Bradypus variegatus. Biotropica, 21, 369.
Haddad N, Brudvig L, Clobert J et al. (2015) Habitat fragmentation and its lasting impact on Earth’s ecosystems. Science Advances, 1, e1500052.
Hayssen V (2010) Bradypus variegatus (Pilosa: Bradypodidae). Mammalian Species, 42, 19-32.
Hirsch A, Chiarello A (2011) The endangered maned sloth Bradypus torquatus of the Brazilian Atlantic forest: a review and update of geographical distribution and habitat preferences. Mammal Review, 42, 35-54.
Lara-Ruiz P, Chiarello A (2005) Life-history traits and sexual dimorphism of the Atlantic forest maned sloth Bradypus torquatus (Xenarthra: Bradypodidae). Journal of Zoology, 267, 63.
Lara-Ruiz P, Chiarello A, Santos F (2008) Extreme population divergence and conservation implications for the rare endangered Atlantic Forest sloth, Bradypus torquatus (Pilosa: Bradypodidae). Biological Conservation, 141, 1332-1342.
Lynch M, Lande R (1998) The critical effective size for a genetically secure population. Animal Conservation, 1, 70-72.
Mendel F (1985) Use of Hands and Feet of Three-Toed Sloths (Bradypus variegatus) during Climbing and Terrestrial Locomotion. Journal of Mammalogy, 66, 359-366.
Mendenhall C, Karp D, Meyer C, Hadly E, Daily G (2014) Predicting biodiversity change and averting collapse in agricultural landscapes. Nature, 509, 213-217.
Mendoza J, Peery M, Gutiérrez G, Herrera G, Pauli J (2014) Resource use by the two-toed sloth (Choloepus hoffmanni) and the three-toed sloth (Bradypus variegatus) differs in a shade-grown agro-ecosystem. Journal of Tropical Ecology, 31, 49-55.
Metzger J, Martensen A, Dixo M, Bernacci L, Ribeiro M, Teixeira A, Pardini R (2009) Time-lag in biological responses to landscape changes in a highly dynamic Atlantic forest region. Biological Conservation, 142, 1166-1177.
Montgomery G, Sunquist M (1975) Impact of Sloths on Neotropical Forest Energy Flow and Nutrient Cycling. Tropical Ecological Systems, 69-98.
Moraes-Barros N, Arteaga M (2015) Genetic diversity in Xenarthra and its relevance to patterns of neotropical biodiversity. Journal of Mammalogy, 96, 690-702.
Moraes-Barros N, Silva J, Miyaki C, Morgante J (2006) Comparative Phylogeography of the Atlantic Forest Endemic Sloth (Bradypus torquatus) and the Widespread Three-toed Sloth (Bradypus variegatus) (Bradypodidae, Xenarthra). Genetica, 126, 189-198.
Moreira D, Leite G, Siqueira M, Coutinho B, Zanon M, Mendes S (2014) The Distributional Ecology of the Maned Sloth: Environmental Influences on Its Distribution and Gaps in Knowledge. PLoS ONE, 9, e110929.
Moritz C (1994) Defining ‘Evolutionarily Significant Units’ for conservation. Trends in Ecology & Evolution, 9, 373-375.
Moritz C (2002) Strategies to Protect Biological Diversity and the Evolutionary Processes That Sustain It. Systematic Biology, 51, 238-254.
Moss W, Pauli J, Gutiérrez G, Young A, Vaughan C, Herrera G, Peery M (2011) Development and characterization of 16 microsatellites for Hoffmann’s two-toed sloth, Choloepus hoffmanni. Conservation Genetics Resources, 3, 625-627.
Moss W, Peery M, Gutiérrez-Espeleta G, Vaughan C, Herrera G, Pauli J (2012) Isolation and characterization of 18 microsatellite markers for the brown-throated three-toed sloth, Bradypus variegatus. Conservation Genetics Resources, 4, 1037-1039.
Nowak R (1999) Walker’s mammals of the world. The Johns Hopkins University Press, Baltimore.
Oliver WLR, Santos FB (1991) Threatened endemic mammals of the Atlantic forest region of South-East Brazil. Jersey Wildlife Preservation Trust, Jersey.
Pauli J, Peery M (2012) Unexpected Strong Polygyny in the Brown-Throated Three-Toed Sloth. PLoS ONE, 7, e51389.
Peakall, R. and Smouse P.E. (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics, 28, 2537-2539.
Peery M, Pauli J (2012) The mating system of a ‘lazy’ mammal, Hoffmann’s two-toed sloth. Animal Behaviour, 84, 555-562.
Primack R (2014) Essentials of conservation biology. Sinauer, Sunderland, Mass.
Pulido-Santacruz P, Renjifo L (2010) Live fences as tools for biodiversity conservation: a study case with birds and plants. Agroforestry Systems, 81, 15-30.
Raymond M. & Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity, 86, 248-249
Reed D, Frankham R (2003) Correlation between Fitness and Genetic Diversity. Conservation Biology, 17, 230-237.
Rousset F. (2008), GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources, 8, 103–106.
Sader S, Joyce A (1988) Deforestation Rates and Trends in Costa Rica, 1940 to 1983. Biotropica, 20, 11.
Santos Júnior J, Santos F, Silveira F (2015) Hitting an Unintended Target: Phylogeography of Bombus brasiliensis Lepeletier, 1836 and the First New Brazilian Bumblebee Species in a Century (Hymenoptera: Apidae). PLOS ONE, 10, e0125847.
Schetino M, Coimbra R, Santos F (2017) Time scaled phylogeography and demography of Bradypus torquatus (Pilosa: Bradypodidae). Global Ecology and Conservation, 11, 224-235.
Sharma S, Dutta T, Maldonado J, Wood T, Panwar H, Seidensticker J (2013) Selection of microsatellite loci for genetic monitoring of sloth bears. Ursus, 24, 164-169.
Silva S, Dávila J, Voirin B, Lopes S, Ferrand N, Moraes-Barros N (2017) The curious case of Bradypus variegatus sloths: populations in threatened habitats are biodiversity components needing protection. Biodiversity and Conservation, 1-18.
Tabarelli M, Pinto L, Silva J, Hirota M, Bedel (2005) Challenges and Opportunities for Biodiversity Conservation in the Brazilian Atlantic Forest. Conservation Biology, 19, 695-700.
Taube E, Keravec J, Vié J, Duplantier J (2008) Reproductive biology and postnatal development in sloths, Bradypus and Choloepus: review with original data from the field (French Guiana) and from captivity. Mammal Review, 31, 173-188.
The IUCN Red List of Threatened Species (2018) Iucnredlist.org.
Vaughan C, Ramírez O, Herrera G, Guries R (2007) Spatial ecology and conservation of two sloth species in a cacao landscape in Limón, Costa Rica. Biodiversity and Conservation, 16, 2293-2310.
Zahawi R, Duran G, Kormann U (2015) Sixty-Seven Years of Land-Use Change in Southern Costa Rica. PLOS ONE, 10, e0143554.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Biology"
Biology is the scientific study of the natural processes of living organisms or life in all its forms. including origin, growth, reproduction, structure, and behaviour and encompasses numerous fields such as botany, zoology, mycology, and microbiology.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




