Messenger RNA Delivery for Tissue Engineering and Regenerative Medicine Applications
Info: 8397 words (34 pages) Dissertation
Published: 24th Feb 2022
Abstract
There is a current thrust towards the development of safe, integration-free approaches to gene therapy for translational purposes. Recent advances in in-vitro mRNA transcription techniques and chemical modifications to curtail immunogenicity, in addition to development of novel and more efficient means of membrane and intracellular delivery, have given mRNA therapeutics an increased advantage over traditional DNA based gene transfer methods. mRNA-derived gene therapy strategies are attractive tools for tissue engineering problems, allowing for precise control over the natural cell signaling mechanisms that may enhance the therapeutic and regenerative functions of stem cells, such as their differentiation and secretome activity, and the reprograming of other cell types. Harmonizing mRNA delivery vehicle interactions with tissue engineering scaffolds could confer an additional level of control over spatial and temporal delivery kinetics and enable targeting of proteins to tissues of interest. While current technologies in synthetic mRNA design and delivery show promise, there is more ground to be covered in terms of characterization of mRNA and vehicle interactions with cells as well as in optimization of microenvironment conditions for delivery. In this review we describe existing methods of mRNA synthesis and modification, their conjugation and encapsulation with nanoparticles, and well as their delivery mechanisms from hydrogel scaffolds. We discuss how novel formulations of polymeric materials with mRNA can offer transformative steps towards controlled regeneration of tissues and organs via genetic manipulation in-vitro and in-vivo.
Keywords: mRNA delivery, gene therapy, cell reprograming, tissue engeering, lipid nanoparticles, hydrogels
1. Introduction
The multidisciplinary field of tissue engineering and regenerative medicine combines technologies derived from chemistry, material sciences, stem cell biology and increasingly more frequently, genetic engineering; all with the ultimate goal of developing functional and clinically useful biological tissue substitutes. In recent years, substantial strides have been made in regenerative medicine through the application of strategies that seek to utilize the therapeutic capacity of stem cells and their inherent regenerative capacity upon tissue injury[1]. In addition to their ability to differentiate into multiple cell lineages in response to signaling factors [2], the stem cell secretome, which comprises a broad array of cytokines, chemokines, and growth factors[3], has been increasingly acknowledged for its role in repair and regeneration, making stem cells uniquely suitable for regenerative applications. Harnessing these functions by directing stem cell fate, function and phenotype, and controlling their natural signaling pathways, can further extend our control over the inherent regenerative capabilities of these systems. In this review, we describe recent examples of how the regenerative capacity of stem cells can be harnessed by the use of nucleic acid therapeutics to either trigger (DNA, messenger RNA) or suppress (small interfering RNA, micro RNA (miRNA), small hairpin RNA (shRNA), antisense oligonucleotides, CRISPR/Cas gene editing) the expression of specific genes and transcription factors, with particular attention to the potential and current use of messenger RNA (mRNA) on various regenerative applications.
mRNA has recently garnered much attention for its potential for gene transfer applications [ref]. This turn of events followed improvements in mRNA synthesis and stability, which have allowed its use in a broad range of applications surpassing the potential of DNA-based applications. While DNA has been explored extensively, it has come up short with respect to clinical translation due to low efficiency [4, 5], risk of insertional mutagenesis [6-8], as well as unpredictable and slow expression kinetics. Therapeutic delivery of mRNA, on the other hand, possesses virtually no risk of genomic integration [9]. Since mRNA does not require nuclear entry (a significant barrier to successful gene transfer using DNA) and needs only to reach the translational machinery in the cytosol for expression of its protein product, it has a higher transfection efficiency than DNA [4] and can transfect non-dividing cells [4, 5]. mRNA transfection is characterized by rapid, transient, and predictable protein expression kinetics allowing greater temporal regulation over protein production. As a consequence, mRNA has been employed in cancer immunotherapy [10-13], vaccine development [14-18], protein replacement therapy [19-21], cellular reprogramming [2, 22-25], and genome engineering [26, 27]. While still in its infancy, mRNA applications in tissue engineering present significant potential where traditional DNA-based therapies have failed due to inherent cellular limitations.
In this scenario, considering that there have been no reviews on this specific theme, this manuscript will summarize the recent advances on the mRNA application in tissue engineering, emphasizing findings that contribute to the better understanding of the molecular mechanisms underlying the regenerating abilities of stem cells and thus potentially impacting the field of regenerative medicine.
2. In Vitro Transcribed (IVT) mRNA
Despite the advent of mRNA delivery in the 1970s by polycations [28] and liposomes [29], the instability (average half-life of 7 hours) [30] and innate immunogenicity [31-36] of mRNA have rendered it unsuitable for therapeutic applications. Significant advances in decoding of mRNA biology have now allowed us to upgrade methods of IVT mRNA synthesis by incorporating various chemical modifications to improve IVT mRNA pharmacokinetics and pharmacology.
mRNA serves as a template for protein synthesis and is made up of four ribonucleoside bases: adenosine, guanosine, cytidine and uridine, linked through a phosphate group. With an average size of approximately 2000 bases, this single stranded molecule can be subdivided into 5 domains – the 7‑methylguanosine cap (m7G) at the 5’ end, 5’ untranslated region (5’ UTR), coding sequence known as the open reading frame (ORF), 3’ untranslated region (3’ UTR), and 3’ poly(A) tail consisting of 50 – 250 adenosine residues (Fig. 1) [37]. Modifications to the 5’ cap, poly(A) tail, and 5’ and 3’ untranslated regions (UTRs) have been explored for modulation of IVT mRNA immunogenicity and stability.

Figure 1 – Key structural domains of mRNA. 7-methylguanosine (m7G) cap, 5’ Untranslated region (UTR), Open reading frame (ORF), 3’ UTR, Poly (A) tail
The 5’ cap and the poly(A) tail are critical in the protection of the transcript. Apart from playing a role in regulation of nuclear export of mRNA, the 5’ 7-methylguanylate cap protects against degradation by exonucleases and facilitates the initiation of translation through the binding of eukaryotic initiation factor 4E (eIF4E) [38]. Common in vitro transcription techniques can result in significant fraction of the caps being introduced in reverse orientation, preventing binding of translation factors [39]. In response to this phenomenon, mRNA is now synthesized with anti-reverse cap analogues (ARCAs), which have greatly enhanced translational efficiency [40, 41]. The poly(A) tail aids in nuclear export of mRNA, promotes translation through interactions with translation initiation factors, and prevents degradation by nucleases [37]. In IVT mRNA, it is elongated either through enzymatic polyadenylation using recombinant poly(A)polymerase or by directly encoding for the poly(A) nucleotides within the DNA template. It has been reported that a poly(A) tail of about 120 nucleotides is necessary for optimal inhibition of mRNA degradation [42].
The 5’ and 3’ UTRs present additional avenues for modifications. The two regions play a role in mRNA translation and stability through the interaction of 5’ UTR with translation machinery and the 3’ UTR serving as a binding site for microRNA (miRNA) and mRNA decay-promoting proteins [43]. A frequently approach for increasing the half-life of IVT mRNA is selecting UTR domains from human mRNAs with higher stability (e.g. human α and β-globin) and incorporating them into the 3’ UTR of IVT mRNA [42].
The mammalian immune system is well-adapted for detecting and eliminating exogenous RNA, recognizing it as a viral or viroid challenge. RNA is identified through pattern recognition receptors (PRRs), Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors (NLRs), possessed by immune cells such as macrophages and dendritic cells [31, 32, 35, 36]. Binding of these receptors triggers an inflammatory cascade, which results in RNA degradation, representing an important consideration for potential application in regenerative medicine. Non-immune cells also detect exogenous RNA through PRRs such as the retinoic acid inducible gene 1 (RIG-1) receptor family to induce innate immune responses [33, 34]. Several chemical base modifications have been examined to restrict immune activation by IVT-mRNA [44-46]. This process involves the substitution of a base (e.g. uridine) with the equivalent modified base (e.g. 5-methyluridine) in the entire sequence (Fig. 2). Since base modification is a naturally-occurring post-transcriptional event in eukaryotes, these chemical modifications do not inhibit the endogenous ribosomal IVT mRNA translation [47, 48], and have represented important improvements on the stability of mRNA for intracellular delivery and their potential use to stimulate the regenerative capacity of stem cells in-vitro and in-vivo. It should be noted that the efficacy of the enhancements conferred by chemical modulation of IVT mRNA seems to depend on the type and method of modification, cell type, and cell differentiation state. Moreover, it is believed that there is still much to be discovered concerning mRNA interactions with the myriad extracellular and intracellular proteins and the full breadth of possible modulations in mRNA engineering is yet to be realized. All of these aspects represent important areas of future study that remain to be explored.
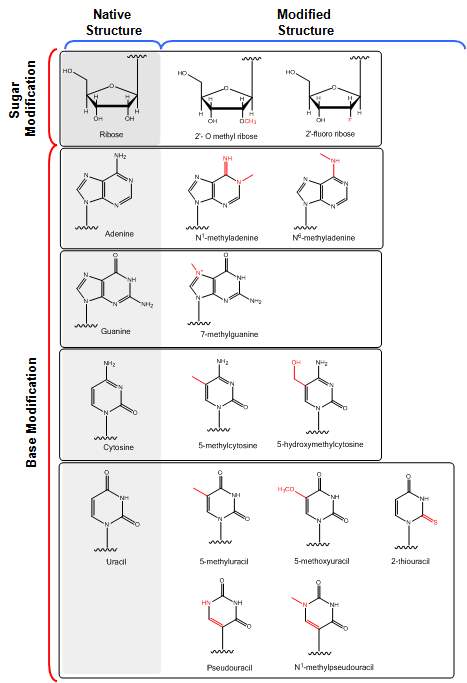
Figure 2. Structural features of different sugar and base modifications to enhance translational efficacy of mRNA with reduced immunogenicity
2.1. mRNA Delivery
The chemical modification of IVT mRNA has greatly improved its efficacy, however, its delivery remains a challenge. It is known that exposure to serum and intracellular nucleases, in addition to immune cells, reduce the half-life of mRNA [31, 33, 49-52]. Moreover, the high molecular weight and negative charge density of mRNA prevents it from effectively entering cells, causing steric inhibition to diffusion and electrostatic repulsion from the anionic cell membrane. Various delivery technologies have been utilized to overcome one or more of these barriers for intracelullar delivery. Here, we will selectively highlight physical and chemical methods that have been reported to circumnavent these barriers. We encourage the reader to refer to a recent review on the topic [Gaurav’s Nature review] for more in-depth details of the described methods.
2.1.1 Physical methods
Physical modes of gene transfer have been developed to achieve direct transfection of cells in vitro and ex vivo. Electroporation is the most popular of these methods demonstrating high efficiency in direct delivery of mRNA into the cytosol [19, 53-56]. This is achieved through the application of electrical pulses to cells in order to permeabilize the cell membrane for the entry of mRNA. Another technique used for cytosolic mRNA delivery utilizes a ‘gene gun’, wherein heavy metal particles (e.g. gold) surface-coated with mRNA are propelled at high velocities into the cell using pressurized gas [57, 58]. Microinjection, yet another physical method, makes use of a microneedle to inject mRNA into individual cells [55, 56]. The low throughput of this technique makes it extremely labor intensive and hence, has only seen sporadic use [55]. Sonoporation, using ultrasound along with microbubbles, has also been used to deliver mRNA to dendritic cells ex vivo [13]. Collectively, these techniques have been used primarily for application in cancer immunotherapy [13, 53], and to lesser extent in protein replacement therapy [19, 58], cellular regrogramming [59, 60] and gene editing [55, 56], where the latter two have especial relevance for the field of tissue engieering. These methods deliver mRNA directly to the cytosol, avoiding the need for endosomal escape and immune activation. The efficacy of physical methods at mRNA transfection is clearly demonstrated by their use in the majority of clinical trials for ex vivo applications. However, these methods remain expensive, highly invasive (for instance, extraction of blood), and require extensive optimization.
2.1.2 Chemical methods
The limitations of physical methods have lead to the development of chemical methods for delivery of nucleic acids. Today, chemical techniques, with their high versatility, low cost and ease of use, have been broadly evaluated for in vitro, ex vivo, and in vivo delivery of nucleic acids. Furthermore, the true advantage lies in their potency to deliver nucleic acids in vivo. Parenteral delivery of nucleic acids is faced with additional challenges such as degradation of nucleic acids by immune cells [31, 36]and serum nucleases [51, 52], and the need for cellular uptake and endosome escape to achieve successful transfection [61, 62]. These problems have been addressed using nanoparticles made using cationic molecules such as polycations, cationic or ionizable lipids and lipid-like molecules (Fig. 3). These cationic molecules bind electrostatically to the anionic phosphates on the mRNA backbone leading to efficient packaging of mRNA into nanoparticles. Evasion of immune recognition is accomplished by shielding the nanoparticles using inert materials, like polyethylene glycol (PEG) [63]. Cellular uptake is achieved through endocytosis following receptor, electrostatic, or hydrophobic binding of nanoparticles to the cell surface followed by internalization [64]. Extensive effort has been put into utilizing ligands (such as transferrin, folic acid, RGD peptide, etc.) towards cell surface receptors to increase specificity and efficiency [61, 64]. A major consideration in design of nanocarriers is their ability to induce endosome escape. Two mechanisms are accepted as potential hypothesis for endosome escape of nanoparticle – proton sponge effect [65] and membrane destabilization [66]. Additional amplification of transfection can be attained through the use of endosomal escape agents such as chloroquine [67], guanabenz [68], endosomal escape peptides (such as melittin, muramyl dipeptide, etc) [67, 69], and leukotriene inhibitors (e.g. MK571, Pranlukast, Zafirlukast) [69].The flexibility in nanoparticle design, size of mRNA, low cost, ease of scale-up, limited toxicity, and low immunogenicity has made chemical modes the preferred choice for mRNA delivery. Selected examples are provided below.
Polycations
Polycations utilize electrostatic interactions to encapsulate nucleic acids ensuring serum stability. The cationic charge of the nanoparticles thus formed also assists in internalization of these polyplexes (complex of polycation and nucleic acid) into cells. One of the first polymers to be investigated for nucleic acid delivery was Poly-L-lysine (PLL) [70, 71] (Fig. 3A). While PLL was proficient at encapsulating nucleotides, it was unable to induce endosomal escape due to its poor buffering capacity and resulting in low efficiency of gene transfer. A new polymer, polyethylenimine (PEI) (Fig. 3A), was later developed to have a high charge density and superior buffering capacity leading to higher efficacy. PEI remains the most explored polymeric delivery material for nucleic acid delivery [67, 72]. Polyaspartate (poly-Asp) (Fig. 3A) is another broadly investigated polycation for nucleotide delivery. Variations of polyaspartatesuch as poly-Asp(DET), poly-Asp(TET), poly-Asp(TEP), poly-Asp(PEH) have been successfully used for mRNA delivery [73, 74]. To further add to the versatility and utility of these polymers, block copolymers of PLL, PEI, and poly-Asp with PEG have been developed to form ‘nanomicelles’ for enhanced in vivo delivery [75-77]. Poly beta amino esters (PBAEs) [78, 79], and polypeptoids [74, 80] are other class of polycations which have evolved in design to improve efficacy and to reduce toxicity.
Lipoplexes and Lipid Nanoparticles (LNPs)
Lipoplexes exploit the mechanism of electrostatic complexation to package nucleic into nanoparticles using cationic lipids (e.g. 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA), N-[1-(2,3-Dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTAP), (Fig. 3B) and/or an accessory neutral lipid (such as 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) (Fig. 3B). Notable cationic lipoplex formulations explored for in vitro, ex vivo and in vivo delivery of mRNA include DOTAP:DOPE, Lipofectamine™, TransIT™, MegaFectin™, and Stemfect™ [20, 72]. Further optimization of formulations containing cationic lipids led to the development of highly modular lipid-based nanocarriers known as lipid nanoparticles (LNPs). Most commonly, lipid nanoparticles are composed of combinations of a cationic lipid, a structural lipid, cholesterol, and PEG-lipid, mixed together in precise ratios. The presence of ‘helper’ lipids (structural lipids, PEG-lipids, and cholesterol) confers stability as well as protection from opsonization and non-specific uptake by mononuclear phagocyte system (MPS) resulting in improved circulation half-life and efficacy [81]. At the cellular level, LNPs are trafficked through the endo-lysosomal pathway with endosomal escape taking place through membrane destabilization initiated by electrostatic interactions between cationic lipids of the LNPs and anionic lipids of the endosomal membrane. Although, the use of cationic lipids in lipoplexes and LNPs offers high transfection in vitro; their toxicity and immunogenicity has led to setbacks for in vivo translation [82-84].
Over the years, better understanding of the necessities in the delivery process have allowed the development of potent synthetic lipids leading to higher efficacy and lower toxicity [85]. The greatest advancements for in vivo delivery of nucleic acids have come in the form of ionizable lipids (e.g. C12-200, TT3, DLin-MC3-DMA, OF-2, Fig. 3B) as replacements for cationic lipids in LNPs. LNPs with ionizable lipids have experienced the most success among nanoparticle gene delivery systems, showing positive results in clinical trials for RNA therapeutics [86]. Patisiran, a LNP containing siRNA against transthyretin (TTR), successfully completed phase 3 clinical trials, making it the first to reach this stage. Several LNP formulations are currently being tested for mRNA delivery in various preclinical and clinical studies including infectious disease vaccinations and cancer immunotherapy [16, 87-89].
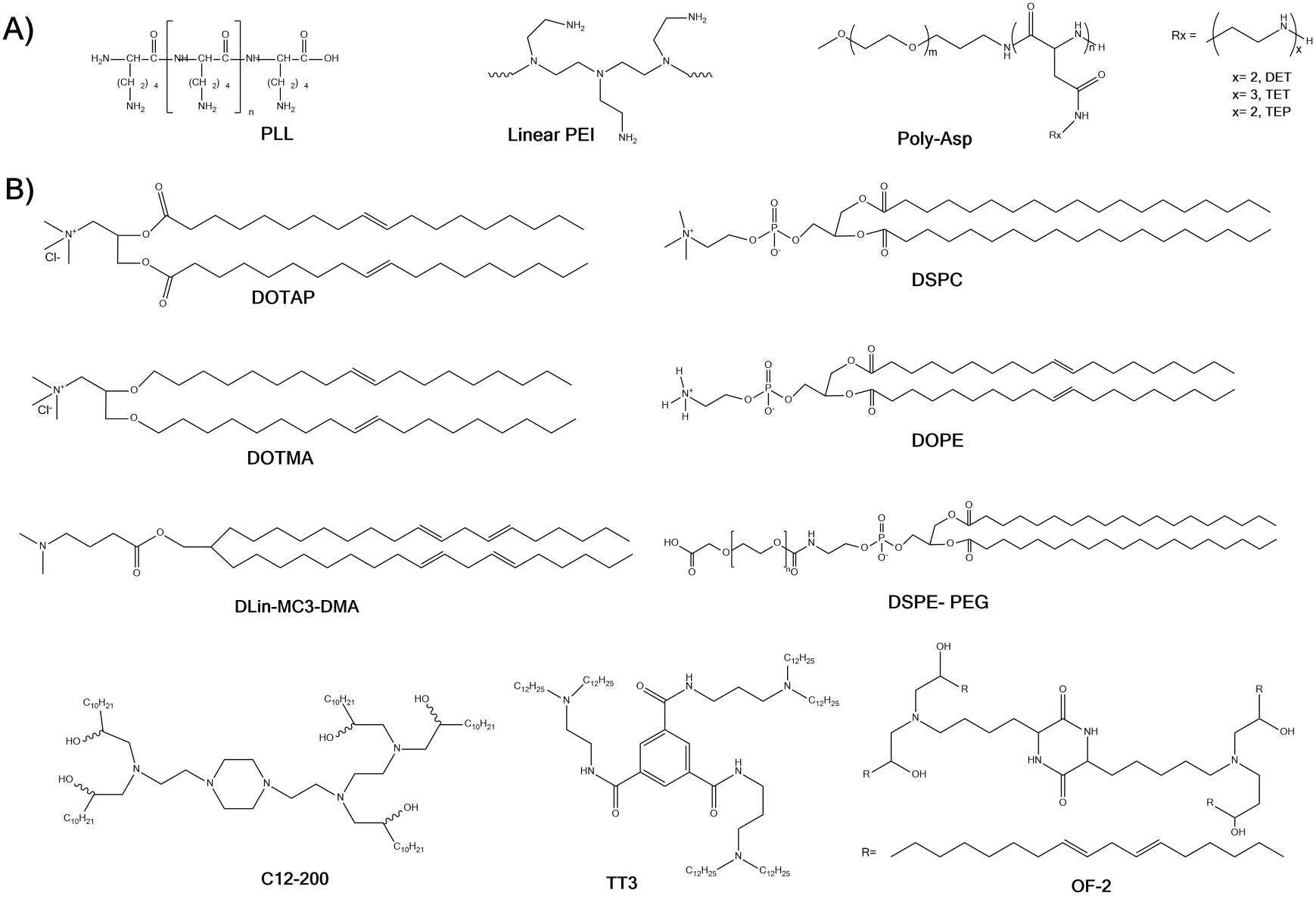
Figure 3. Structures of A) polycations and B) lipid and lipid-like materials developed for in vivo mRNA delivery.
3. Hydrogel scaffolds in mRNA delivery
A noteworthy aspect that requires especial attention regarding the use of mRNA for tissue engineering is that, different from RNAi strategies, where nucleic acids are mostly used for down regulation of intracellular mechanisms, mRNA use in tissue engineering is primarily intended to promote tissue formation or certain aspects of cellular response, such as differentiation and protein secretion. However, given the 3D architectural complexity of tissues in the body, such an mRNA-guided response ideally needs to be controlled in three dimensions and in a site-specific manner, thus giving rise to 3D multi-typic tissues with spatially defined functions and organization, such as those in the body. Therefore, local and sustained delivery of mRNA is a challenge that becomes particularly relevant for tissue engineering strategies. Effectively, this means that much of the optimization that has been performed in the field of mRNA complexation, as well as bolus intravascular mRNA delivery trhough nanoparticles, both which represent the bulk of the literature in the field, become only a stepping stone for future developments on controllable biomaterial “depots” that are compatible with such delivery vehicles. Moreover, in addition to being compatible with these nanodelivery vehicles, such biomaterials need to be compatible with the cells that are being used for new tissue formation. Importantly, the far majority of tissue engineering strategies in the past decade have used cell-laden biomaterials that attempt to replicate the native microenvironment that cells are exposed to in the body, which makes these challenges especially more complicated.
For the most part, challenges associated with design and fabrication of complex tissues in the lab, a field that has emerged under the broad definition of “biofabrication” or “biomanufacturing”, have used various types of scaffold materials. These have generally been based either on solid slow-degrading or non-degrading systems (i.e. bioceramics, metallic implants, etc.), or on biodegradable polymeric materials, out of which hydrogels have been the ones with far greater attention in tissue engineering. Given the inherent compatibility of hydrogel scaffolds with mRNA nanoparticles, lipoplexes and polyplexes, this review will cover primarily aspects that relate to polymer hydrogels. The reader is encouraged to refer to recent comprehensive reviews on other materials for non-viral intracellular delivery of mRNA in recent publications [Ref].
Hydrogels are three-dimensional networks of polymers that exhibit the ability to swell and retain large amounts of water within their structure. We have recently published a comprehensive review of their recent use in tissue engineering for in various applications, and the reader is encouraged to refer to this publication for details [Adv Mater Rev]. Natural hydrogels are engineered based on naturally occurring proteins (e.g., collagen, gelatin, and fibrin), and polysaccharides (e.g., alginate chitosan, hyaluronic acid, dextran); whereas synthetic hydrogels are made from synthetic polymers, such as poly (acrylic acid) (PAA), poly (ethylene glycol) (PEG), poly (vinyl alcohol) (PVA), polyacrylamide (PAAm), and other polypeptides [90]. Much of the excitement surrounding polymer hydrogels in tissue engineering stem from their ability to be tailored for specific applications, which can be achieved by carefully tuning their biodegradability, physical and mechanical properties, injectability, and microstructure, either by controlling their polymer to water ratio, or their degree of polymerization upon gelation [Ref]. Additionally, hydrogels can be engineered to be responsive to specific stimuli, such as enzymatic activity [ref], pH [ref], temperature [ref], light [ref] and electricity [ref]. These different components of customization render hydrogels with improved pharmacokinetic and pharmacodynamic properties, and added to their inherent capacity for overcoming problems such as off-target nucleic acid accumulation [ref]. For these reasons hydrogels represent a highly desirable material for mRNA delivery in tissue engineering.
Similar to other aspects of genetic manipulation discussed above, DNA and RNAi delivery using hydrogels have dominated the field for quite some time [ref], and the use or mRNA for regeneration has only recently received more attention. Of note, like in all other nucleic acid-based therapies, mRNA is a large polyanion that does not readily cross non-polar cellular and tissue barriers. Therefore, the potential interactions occurring between the mRNA delivery vehicles with hydrogels is a point that requires careful attention. In addition to their composition, crosslinking density and delivery method (injectability, in-situ polymerization, etc.), which are challenges that are somewhat common to other drug delivery strategies, the electrostatic interactions between hydrogels with mRNA is a factor that needs to be carefully considered, both when mRNA is directly included in the gel network (naked mRNA), or loaded into nanoparticles.
Given the polyanionic character of mRNA, and cationic property of most nanoparticles used for their delivery, hydrogels that are electrostatically inert have been proposed as improved materials for mRNA based tissue engineering, these include priamrely PEG based hydrogels and dextran [62, 85 AHM]. Cationic hydrogels, however, such as gels composed of Polyethylenimine (PEI), chitosan, polyamidoamine, poly β-amino-esters (PBAEs) and poly L-lysine, have been shown to confer advantages to nucleic acid delivery, including mRNA [AHM]. This is because cationic polymers not only have the ability to self assemble into hydrogels themselves [59, 88 AHM], but also because their positive charge distribution enables sustained release of sequestered nucleic acid cargo due to the net negative charge of mRNA and other nucleic acid nanoparticles. An important disadvantage of these materials for tissue engineering, however, is that cationic polymers have been known to activate to apoptotic signaling pathways [92] and to induce formation of nanopores on the cell membrane [93, 94], which can lead to cytotoxic effects despite the enhanced transfection outcomes. Moreover, hydrogels derived from these polymers by themselves lack cell adhesive ligands (RGD, MMP) that are known to regulate important mechanisms of cell response and differentiation [Ref]. Therefore, future work should seek to design and develop less cytotoxic cationic hydrogels that are compatible with cell loading, existing biofabrication methods, while still offering favorable electrostatic interactions with polyplexes, lipoplexes and naked nucleic acids. Current research has paid particular attention to the development and modification of PEI with various additives to improve their cytotoxicity [95-99], although modification of PEI and other cationic hydrogels for improved mRNA delivery, specifically, has remained relatively stagnant in recent years. Lastly, while cationic polymers have been linked to a more sustained delivery of genetic cargo, negatively charged nanoparticles or mRNA complexes may be more readily repelled in anionic hydrogels [AHM], which is likely to enhanced their immediate delivery in areas where fast-release is desired, such as for rapid vascularization of engineered tissues or wound healing.
Despite being an area of certain interest in future, relatively few classes of materials have been evaluated as hydrogels for mRNA delivery. The far majority of the work performed using hydrogels has been performed on RNAi strategies, and the reader is encouraged to refer to a recent review on the use of hydrogels for RNAi therapies [AHM]. Of relevance, it has been demonstrated that nucleic acid delivery in 2D and 3D hydrogels can activate sharply different signaling pathways, although the targets may be similar [Segura Integrative Biol 2013]. Krebs et al compared the ability of the released nucleic acid to knock down the expression of GFP in cells that constitutively express this protein among three different types of cell-laden hydrogel scaffolds: calcium crosslinked alginate, photocrosslinked alginate, and collagen. The results showed that alginate hydrogels resulted in rapid release of nucleic acid cargo over one week, whereas collagen had more prolonged and controlled release over a period of two weeks; the differences which were postulated to result from the electrostatic interactions between the gel matrix and the RNA cargo. Cell-laden PEI/dextran hydrogels that had complete degradation over 17 days demonstrated 80% efficiency in GFP expression knockdown when siRNA was released from the hydrogel matrix [85 AHM]. A subsequent report utilizing siRNA for osteogenic differentiation in cell-laden PEG hydrogels utilized a thiol-achrylate reaction for in-situ hydrogel gelation, showing a marked increase of cell differentiation upon release of RNA interfering molecules in a sustained and controlled manner over the course of 3-6 weeks [82 AHM]. Spatially controlled release of nucleic acid in hydrogels was also shown in methacrylated dextran hydrogels loaded with RNA/PEI polyplexes using a syringe pump, demonstrating that localized and site-specific cell response can be achieved through the controlled gradient formation of the hydrogel scaffold. Recent preliminary experiments from our group using gelatin methacryloyl hydrogels photopolymerized using a visible LED light and loaded with stem cells from teeth showed effective intracellular delivery of mRNA for expression of luciferase, where the mRNA release was directly proportional to the hydrogel concentration over a period of 48 hours (Fig X). An important observation that requires attention is that multiple groups have reported that directly adapting DNA or siRNA methods for mRNA delivery can be ineffective [PNAS], which encourages future studies on the specific use of of hydrogels for mRNA delivery in tissue engineering.
In summary, the vast amount of tools available for the development and tuning of hydrogel scaffolds ha has been generated in the past two decades in the field of tissue engineering will be importnat to develop improved methods for mRNA delivery. Although the field of genetic manipulation for tissue engineering has been largely skewed towards the use of DNA delivery and RNAi in recent years, there is growing evidence of the ability of mRNA to address important challenges in regenerative medicine. The ideal combination of scaffold-guided cell response with specific mRNA delivery vehicles remains to be determined. However, this is certainly an area that deserves to be explored for future regenerative applications.
4. mRNA applications in tissue engineering
4.1 mRNA Delivery for Induced Pluripotent Stem Cells (iPSCs) Reprogramming
The ability to derive pluripotent stem cells from somatic cells through the induction of the Yamanaka reprogramming factors (Oct-4, SOX-2, KLF-4 and c-Myc)[91, 92] has uncovered an autologous source of stem cells for regenerative medicine, without the ethical issues associated with human embryonic stem cells, or the logistics challenges associated with primary mesenchymal stem cell isolation. Cell reprogramming was originally achieved through viral transduction and integration of these reprogramming factors into the genome of host cells. This approach, however, precludes their use for therapeutic applications. Since continuous ectopic expression of these reprogramming factors is both unnecessary [93, 94] and undesirable [95, 96], transient transfection strategies involving plasmid vectors [97, 98], adenovirus [99], DNA transposons[100-102] or excisable lentiviral vectors[103], have been employed to induce pluripotency, which have considerably reduced the risk of genomic modification or insertational mutagenesis. However, a consequent reduction in transfection efficiency and levels of expression has been observed using these methods, highlighting an area where mRNA transfection might be of interest. Although, the Sendai virus, which is a non-integrating vector, provided a high degree of transfection and cytosolic expression, it required elaborate treatment steps to expunge the virus from the cells [103-107]. Other studies have used recombinant proteins, modified to enhance cell penetration[108-110], or nanoparticles[111] to deliver reprogramming transcription factors into cells. Again, the cost of production and purification of recombinant proteins does not justify the low expression and pluripotency induction obtained by these methods. Therefore, mRNA intracellular delivery for somatic cell reprogramming has been an emerging area of interest in the tissue engineering community, since, in principle, it should enable more straightforward protein translation and reprogramming efficiency.
Synthetic mRNA was first used by Warren et. al. to direct reprogramming by inducing transgene expression of Oct-4, SOX-2, KLF-4, c-Myc and Lin-28 in fibroblasts. Phosphatase treatment of mRNA and substitution of ribonucleoside bases 5-methylcytidine and pseudouridine for cytidine and uridine respectively enabled relatively high doses of transfection without generating host immune response and cytotoxicity, and cells transfected with modified mRNA showed high dose dependent transgene expression[112]. The use of feeder cells and supplementation of medium with the interferon inhibitor B18R further diminished toxic effects of the transfection process. The protocol required periodic, multiple transfections for sustained expression of reprogramming factors over the 2 week duration necessary to induce pluripotency owing to the transient nature of mRNA translation kinetics and degradation, with transgene expression peaking at 24 hours [113]. While the time and labor intensive nature of this method may be considered a disadvantage – and can be remedied through used of scaffold mediated mRNA/nanoparticle release kinetics, as discussed above – the process also renders flexibility in the stoichiometric combination and temporal expression of the transcription factors[114], which is expedient. In fact, a dose ramping regimen developed to adjust mRNA dose to cell proliferation rates resulted in a significant reduction in apoptosis caused by stress during standard reprogramming protocols [cite thesis]. A subsequent publication by Warren et. al. also described the use of a variant of Oct4 with a myoD transactivation domain that facilitated more efficient and faster induction of pluripotency, in addition to other improvements to culture conditions such as 5% O2, addition of valproic acid to medium [113], and a more recent study that incorporated two more reprogramming factors (Lin28 and Nanog), in addition to the Oct4 variant, saw additional improvement in reprogramming under hypoxic conditions[115]. Other combinations of reprogramming factor with unmodified mRNA and in the absence of interferon inhibitors have also since been employed to induce pluripotency in fibroblasts [116], but mRNA reprogramming has yet to be applied to other cell types and different media conditions. A recent comparative study conducted on the efficacy of mRNA reprogramming showed highest efficiency among non-integrating reprogramming methods with epigenetics, homogenity and pluripotency markers comparable to lenti-derived iPSCs although successful mRNA reprogramming could not be achieved in blood cells[117].
As the field of tissue engineering becomes more reliant on the use of reprogrammed cells for cell therapy and regenerative medicine, improvements in the methods of cell reprogramming via nucleic acid delivery are likely to become more relevant. And although both the reprogramming efficiency and mRNA delivery still need to be significantly improved for ultimate clinical use, the efforts expended on phase I/II clinical trials using naked, chemically modified or protamine-complexed mRNA show great promise for the use of mRNA-guided reprogramming in future regenerative efforts [24-26 PNAS refs].
4.2 mRNA Directed Cell Differentiation
Stem cell differentiation occurs during development of an organism to produce new specialized cells, and also in adults to replace cells that are lost [118]. Increased understanding of the specific signaling mechanisms that control stem cell fate and lineage commitment through high throughput DNA and proteomic analyses of the changes in gene expression during self renewal and differentiation of stem cells has enabled directed differentiation of stem cells, particularly in the absence of tissue specific cues. Extracellular cytokine and growth factor defined protocols for differentiation are largely inefficient and present a high degree of variability. For many years now, both viral and non-viral[119] gene delivery methods have been used for transfer of lineage specifying transcription factors in order to direct differentiation of stem cells to desired lineages in a controlled manner[ref]. Again, these methods entail varying levels of genomic integration, risk of immunogenic response and possibly tumorogenesis, thus limiting their translational appeal. The aforementioned progress in reprogramming fibroblasts to pluripotent states through synthetic modified mRNA derived transgene expression of transcription factors suggests that this strategy may be feasibly applied to direct differentiation in stem cells. Following this line of thinking, a proof of concept study conducted by Warren et. al., achieved terminal myogenic differentiation of mRNA induced pluripotent stem cells (iPSCs) to myoblasts by subsequent transfection with modified mRNA encoding MyoD[112] in time scales comparable to other transfection methods[120]. Two other studies have reported transdifferentiation of fibroblasts to myoblasts by mRNA driven exogenous expression of MyoD1 in murine and human models opening up the possibility of other phenotypic conversions using this method[115], [ref 153 from thesis]
Transgene expression of lineage specifying transcription factors to direct differentiation in stem cells or promoting transdifferentiation of somatic cells[ref] through ectopic expression of molecular switches, eliminates heterogeneity caused by defined media formulations, allowing reproducibility of differentiation protocols for regenerative applications with quantitative control over expression levels.
4.3 mRNA delivery to control stem cell secretome
In addition to their multipotent differentiation potential, the regenerative capacity of stem cells is attributed to their robust paracrine response to tissue injury. In particular, the MSC secretome has been found to be pivotal in modulating immune/inflammatory response, regulating angiogenesis and maintaining a cytoprotective effect on local cells through both soluble factors and horizontal gene transfer by extracellular microvesicles and exosomes, in response to stress derived signaling[3]. High throughput analyses using Liquid Chromatography with Tandem Mass Spectrometry Detection (LC-MS/MS), antibody arrays and bioinformatics have identified over 200 proteins as part of the MSC secretome[121]. While pre-conditioning and molecular stimulation protocols have been developed to stimulate the secretory function of stem cells for therapeutic applications, genetic manipulation through overexpression of selected genes such as Akt, and its target genes IGF-1, VEGF, GATA-4 and SDF-1 produced a sustained secretory response.[122-126] Other studies have identified regulators of the stem cell secretome such as calcium/calmodulin-dependent protein kinase kinase-1 (CAMKK1)[127] and the activated form of HIF1[128], over expression of which resulted in stimulation of the secretory function in MSCs through the Akt/PI3K pathway. There is also evidence for the involvement of PI3K, ERK1/2, p38 MAPK and JAK/STAT signaling cascades in the regulation of stem cell secretory actions[129-131]. While controlled genetic manipulation of the stem cell secretory function would require a deeper mechanistic understanding of the specific function of each of these transcription factors and the response they generate, currently, overexpression of individual growth factors such as VEGF, PDGFR, BDNF, with well characterized roles in angionesis and neuroprotection in transplanted stem cells have found use in recovery and regeneration of tissues, particularly for cardiovascular and neuroregenerative applications [REFs]. These strategies, in addition to the concerns associated with traditional gene transfer methods, may warrant undesirable side effects as a result of prolonged expression of paracrine factors [132, 133], which can be avoided by mRNA driven therapeutics.
VEGF-A, one of the crucial factors in angiogenic signaling, was stably and transiently expressed at physiologically relevant doses in multipotent human embryonic stem cell–derived heart progenitors using chemically modified mRNA encoding VEGF-A and stable accumulation of secreted protein occurred for upto 3 days after transfection. While local administration of modified VEGF mRNA after myocardial infarct showed promise in preventing scar formation and engendering regenerative effects through targeted, localized signaling[24], both secretory therapeutic functions as well as vasculogenic lineage specification to an endothelial phenotype was achieved in VEGF engineered progenitor cells in vitro and in vivo[134] evidencing complete functional potency of the translated protein. Co-transfection of multiple signaling factors of P-selectin glycoprotein ligand-1 (PSGL-1), Sialyl-Lewisx (SLeX) and Interleukin-10 (IL-10) in MSCs was successfully performed which provoked an immunomodulatory response in vitro[135] and in vivo[136] as yet another example of enhanced and targeted therapeutic function of mRNA engineered stem cells.
Another interesting application has been proposed for stem cells in regenerative medicine involving the use of microvesicles and exosomes shed by these stem cells as vehicles of genetic material, following their intrinsic function of transporting proteins beyond cell membranes [137, 138]. Their innate stability against enzymatic degradation, long half-life and membrane penetrability add to their appeal and can potentiate cell-free, tissue specific gene modification strategies for regenerative applications[139]. Microvesicles can be modified to be enriched with specific proteins for transfer to target cells by genetic engineering host stem cells for overexpression of these proteins, as a therapeutic or regenerative strategy that surpasses delivery and protein stability limitations faced by other drug or nucleic acid delivery methods[140, 141]. However, more extensive biological characterization of microvesicles will need to be performed before they can be adopted for translational purposes in regenerative medicine.
5. Challenges and perspectives
The field of tissue engineering has evolved dramatically since its inception in the late 90s. Current methods of biofabrication for direct engineering of tissues and organs have represented important strides towards clinical application. Similarly, the developments of advanced in-vitro tools for drug discovery and disease models, such as integrated organs-on-a-chip [Ref], and organoid engineering [Ref] have drastically changed the landscape of research on regenerative medicine. The development of improved methods of genetic manipulation of stem cells for regenerative applications certainly represents another facet of the next-generation of tissue engineering methods. Borrowing tools from basic biology and metabolomics [Ref], proteomics [Ref] and materials sciences/materiomics [Ref], we envision that the generation of libraries [Ref] of polymers will enable the expedited determination of the ideal combinations of nanoparticle carriers, RNA cargos, and hydrogel scaffolds for controlled cell response. Existing microarray technologies may be of interest in the development of these databases of materials and biological systems to better determine how they interact with one another for ultimate use.
Another area that is likely to receive increasing attention is the integration of genetic modifiers, such as mRNA, RNAi, and DNA, in controllable materials for biomanufacturing. As we hinted above, the complexity of native tissues in the body give rise to the need for biofabrication methods that can precisely control the position, size and morphology of each individual component making up complex tissues and organs. Therefore, it is unlikely that this is a situation where a single nanoparticle/mRNA will be able to allow for regeneration of an entire compex tissue or organ. Most likely the combination of multiple nucleic acids and cell-laden hydrogels engineered trhough biofabrication methodswill bring us one step closer to the controlled formation of functional tissues and organs. In this regard, manufacturing tools such as 3D bioprinting [Ref], hydrogel microfabrication [Ref] and microfluidics [Ref] will become increasingly important, especially when utilized with the right cells and scaffold materials. Moreover, the ability of controlling the genetic composition of individual cells in-vitro and in-vivo will open up a range of possibilities that remain poorly explored, including in-situ reprogramming of stem cells after cell-laden hydrogel implantation, in-situ genetic manipulation of delivered cells as a means of immunomodulation, control of genetic vaccines, and other areas that are still in their infancy.
6. Conclusion
A new generation of synthetic nucleic acids are finding favor for gene modification in a wide variety of applications by enabling adjustable transgene expression without risk of insertational mutagenesis or genome modification. While innovative approaches to in vitro transcription and modifications to mRNA chemistry as well as delivery mechanisms have curtailed immunogenicity and improved transfection efficiencies, additional endeavors involving a combination of physical, chemical, biological and engineering techniques will be required to integrate mRNA mediated gene transfer with tissue engineering for regenerative medicine.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Biomedical Science"
Biomedical Science focuses on how cells, organs and systems function in the human body and underpins much of modern medicine. Biomedical Science applies parts of natural and/or formal sciences to help develop advances in healthcare.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




