Natural Language Processing for Improving the Sensitivity and Specificity of Clinical Decision Support
Info: 6332 words (25 pages) Dissertation
Published: 9th Dec 2019
Tagged: Health
Natural language processing for improving the sensitivity and specificity of clinical decision support SYSTEM AND reduce alert fatigue
TABLE OF CONTENT
| PAGE NO | CONTENT |
| 2 | ABSTRACT |
| 3 | BACKGROUND AND DESCRIPTION |
| 5 | LITERATURE REVIEW |
| 10 | METHOD |
| 13 | RESULT AND DISCUSSION |
| 19 | CONCLUSION |
| 21 | REFERENCE |
ABSTRACT:
Over the past ten years, the healthcare system in the United States has drastically evolved from a paper-based to an electronic based system. The electronic system stores enormous amount of data, formally called as Patient records, which are then processed and used by caregivers to make clinically informed decisions. The process of making the clinically informed decision is delivered by clinical decision support system, popularly known as CDS. The format and the capacity of data processed at a time is huge. Over the years, with the introduction of medical format standards such as FHIR and HL7, CDS has gotten around the practice of processing standardized data but also has developed some daunting challenges such as alert fatigue. The most neglected part of the patient records is the non-standardized data, stored as physician and registered nurses (RN) notes. Natural language processing (NLP) promises to break grounds in this aspect. The ability of the NLP concepts to process the non-structured notes is proving to make the CDS more sensitive and specific to the data. This paper is written in collaboration with Brigham and Women’s hospital as a part of the project CONCERN (Collins, 2018). In this paper, using literature review, we are going to identify NLP concepts that would process unstructured caregivers notes and do an exploratory analysis that will help to understand how it improves the sensitivity and specificity of CDS. The caregiver’s notes will be processed to identify onset of sepsis in patients. The results of the coding process will be used to demonstrate the NLP concept, suggest the future steps to integrate into the CDS, and hence possibly reduce CDS challenges such as alert fatigue.
BACKGROUND AND DESCRIPTION:
Clinical decision support system (CDS) is a tool used by care deliverers to effectively cater the needs of the patients by leveraging their health information (Bates et al., 2003). The introduction of clinical decision support system happened in 1959 as a stand-alone system, and evolved as service based system by 2005 (Wright & Sittig, 2008). The invasion of informatics in medicine began with Ledley and Lusted, as Ledley invented CT scanner and Lusted pioneered in Medical decision making (Ledley & Lusted, 1959)(Sittig, Ash, & Ledley, 2006). Fast forward to 2007,CDS had already evolved from a Phase 1:Stand-alone systems to Phase 4: Service models due to countless number of research and integration of machine learning and Artificial intelligence techniques (Wright & Sittig, 2008). As CDS evolved in past few years, challenges hindering the delivery of care has also increased. Some of these challenges are improving human-computer interface, publicize best CDS practices, use of free text in CDS (Sittig et al., 2008). One of the great challenges in CDS today is the occurrence of alert fatigue. As the alerts are more than frequently deployed to the caregivers, they tend to be ignored. Historically, it has been observed that the alerts were the cause of potential drug interactions (Kesselheim, Cresswell, Phansalkar, Bates, & Sheikh, 2011).In most of the cases, these alerts are ignored because they are “clinically irrelevant, goes against patient tolerance or contradicts the providers intention to monitor the patient ”(McCoy, Thomas, Krousel-Wood, & Sittig, 2014). A systematic literature review done on the quality of decision support in computerized provider order entry strongly suggests that the CDS systems must adapt to optimizing the systems with knowledge base to possibly reduce the irrelevant alerts (Carli, Fahrni, Bonnabry, & Lovis, 2018).
Research study says that the physician and RN notes contain vital information about onset of various diseases such as cardiac arrest and sepsis (Ohno-Machado, 2011). In an attempt to discover peripheral arterial disease cases in Mayo Clinic, researchers found that when analyzing radiology notes using predictive algorithms, the system became more specific and sensitive compared to just using structured data (Savova, Fan, et al., 2010). Not only this, as the CDS system become more specific and sensitive to the incoming data, it proves to reduce the number of interruptive alerts (Coleman et al., 2013).Studies suggest that as we integrate more specific algorithms with our CDS, it can drastically improve the delivery of patient risk specifying alerts and hence enhance the quality of care (Coleman et al., 2013).
Sepsis and Clinical decision support system:
Sepsis is one of the most common and fatal medical conditions associated with many patient deaths in the United States of America (Angus et al., 2001). Every year 50% of the patient death is associated to Sepsis(Apostolova & Velez, 2018). Most of these deaths are speculated to have occurred because the CDS system fails to identify the onset of the infection in a patient (Apostolova & Velez, 2018). Hence, an intervention to curb the infection by early detection is needed. In a study done at Rush University Medical Center, Chicago, IL, an attempt to standardize the early recognition and treatment workflow to reduce both mortality and cost of healthcare showed that the evidence based care facilitated improvement in the early recognition of sepsis (Dale, Barbee James, Casey, & Stein, 2016). Lab data and Vital data have been used to detect sepsis using SIRS and SOFA scores (Marik & Taeb, 2017). One of the most recent prediction model that has said to outrun both SIRS and SOFA scores is Insight, which is machine learning based algorithm (Mao et al., 2018).
A number of algorithms have been created to identify Sepsis using combination unstructured notes and structured data. Utilizing nursing notes, in combination structured health data, to create an automated alert system achieved an high F1-score and showed promising results by detecting patients at risk of sepsis (Apostolova & Velez, 2018). Another similar study done to create an automated trigger for Sepsis in emergency showed massive success when the unstructured data was used in combination with structured data. The data was fed into a Natural language processing (NLP) algorithm and then a predictive model was built to accurately predict the cases of Sepsis from the data (Horng et al., 2017). The results of this research showed that when the NLP algorithms were introduced into the predictive model, the system became more sensitive and specific to detecting positive cases of sepsis. In the following research we will use a different set of health data together with notes data to explore the possibilities with NLP and try to identify cases of sepsis.
Goal of this paper:
With the help of Natural language Processing (NLP) we will explore how we can utilize the unstructured care givers notes to improve the sensitivity and specificity of the CDS towards detecting cases of patients at risk to sepsis. NLP concepts will be identified using literature review and an exploratory analysis will be done to understand how these concepts process the unstructured notes. The results obtained will be used to suggest further steps for building a more specific predictive model and reduce alert fatigue.
Literature review:
Natural Language processing and clinical decision support system:
A service based CDS provide efficient support due to the involvement of machine-learning and data mining techniques (Wright & Sittig, 2008). In the recent years, CDS based on Natural language processing (NLP) has shown major breakthrough in care delivery. A number of pre-existing NLP based system such as cTakes are said to show massive success in understanding clinical notes and eventually delivering timely care (Savova, Masanz, et al., 2010). NLP is said to have the potential to drastically improve the three major components of CDS – clinical knowledge, interventions , and decision rules (Demner-Fushman, Chapman, & McDonald, 2009).
NLP is a branch of computational science that processes free text to build different types of systems (Joshi, 1991). There are two types of CDS integrated with NLP – Active and Passive, facilitating functions such as sending out alerts, reminders, coding, and monitoring (Demner-Fushman et al., 2009). NLP has also shown a massive success in creation of task specific CDS system. In one such attempt, radiology notes were used to process the NLP concept based CDS, to detect peripheral artery diseases. The NLP based system, when compared to a baseline system, proved to have accuracy agreement and gold standard of 0.930 (Savova, Fan, et al., 2010). Not only does NLP increases the efficiency of CDS, it reduces the vigorous task of manual labor like data abstraction by 90% (Carrell et al., 2014).
The NLP system or module can be integrated with CDS in various ways. The aim of a NLP or multiple NLP modules involvement in a CDS system is to satisfy either of the three relationships; integration with CDS to achieve a general cohesiveness in terms of data, govern CDS to provide knowledge driven support, and/or specialization of the CDS for a specific task (Demner-Fushman et al., 2009). Keeping this relationship in mind there are two types of NLP systems- Coupled NLP system and Integrated NLP system. Coupled NLP system (Figure 1) is the type which is task specific and works only when evoked by the task centered data. Integrated NLP system (Figure 2) is the type which in general processes data irrespective of a specific task assigned (Demner-Fushman et al., 2009). The systems are made up on the building blocks of NLP, namely, Text preprocessing and Entity recognition (Demner-Fushman et al., 2009).
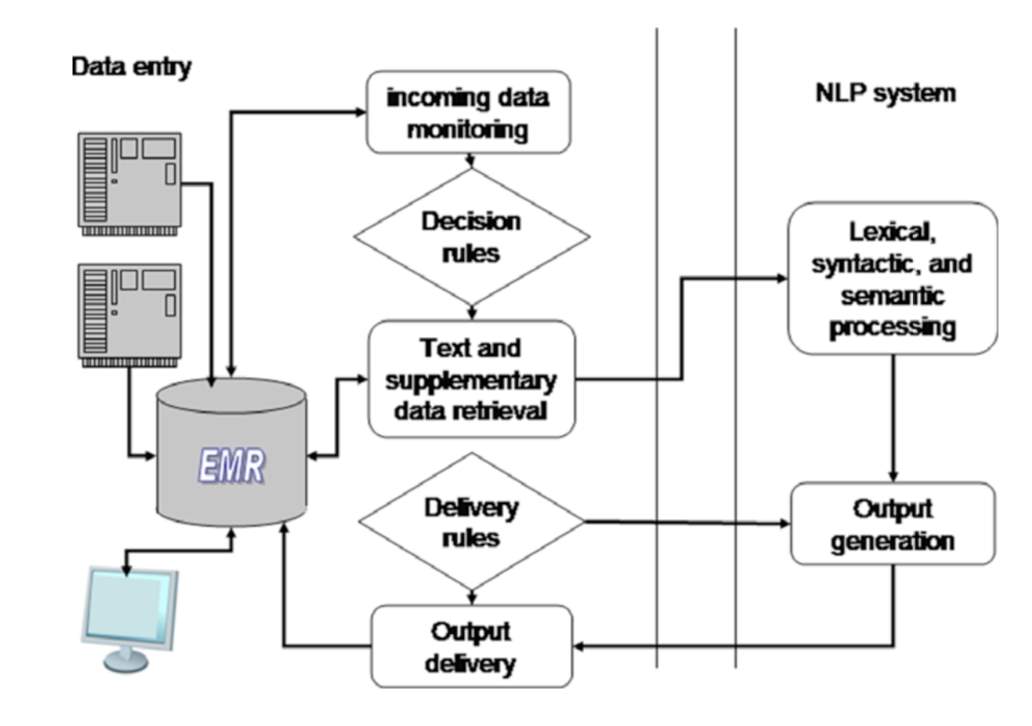
Figure 1: A coupled NLP system (Demner-Fushman et al., 2009)
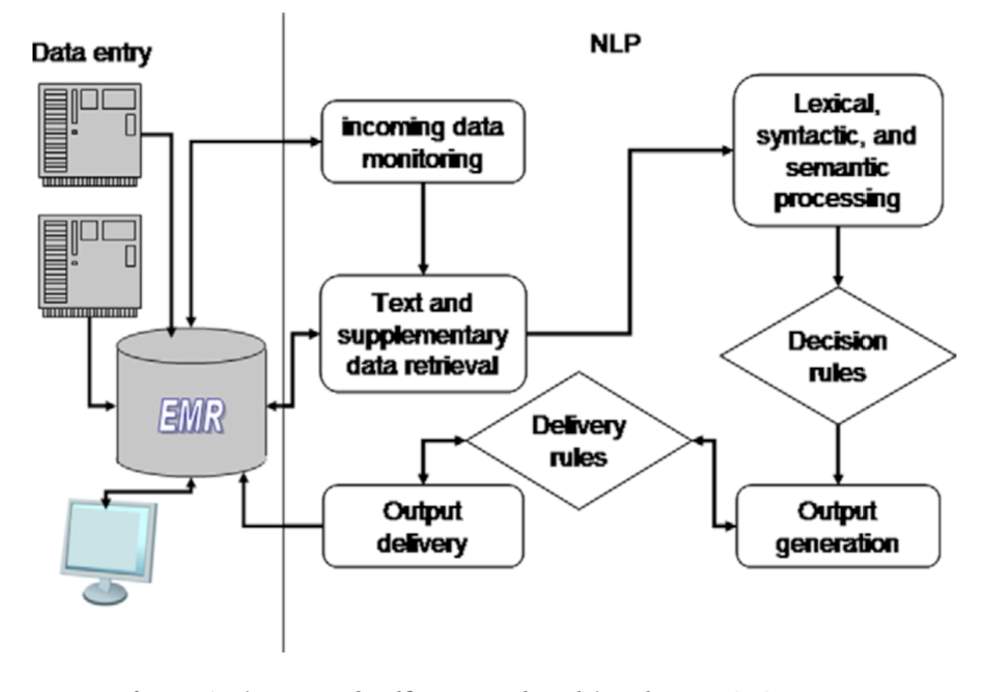
Figure 2 : An integrated NLP-CDS system (Demner-Fushman et al., 2009)
Generally, the basic approach of building an NLP based system is driven by understanding various theories, constructing knowledge bases and application to a reference standard training corpus (Carrell et al., 2014). As the name suggests, NLP takes the data through a number of processing steps before it is finally ready to be used by any system. The unstructured data goes through a number of processes which includes – text parsing, text representation, Tagging, and Modelling or Pattern Mining (Sarkar, 2018). In a more simpler term, NLP would interpret the possible meaning of the sentence and identify clinical concepts in the unstructured dataset using the rules of linguistics (Liao et al., 2015). As the process considers the identified concepts and tags each word in the text to the concepts, the data extracted will be the result of a more sensitive search. The CDS system becomes more specific as other data processing techniques, such as Machine learning algorithms and statistical analysis, are incorporated with NLP’s data extraction (Liao et al., 2015) .
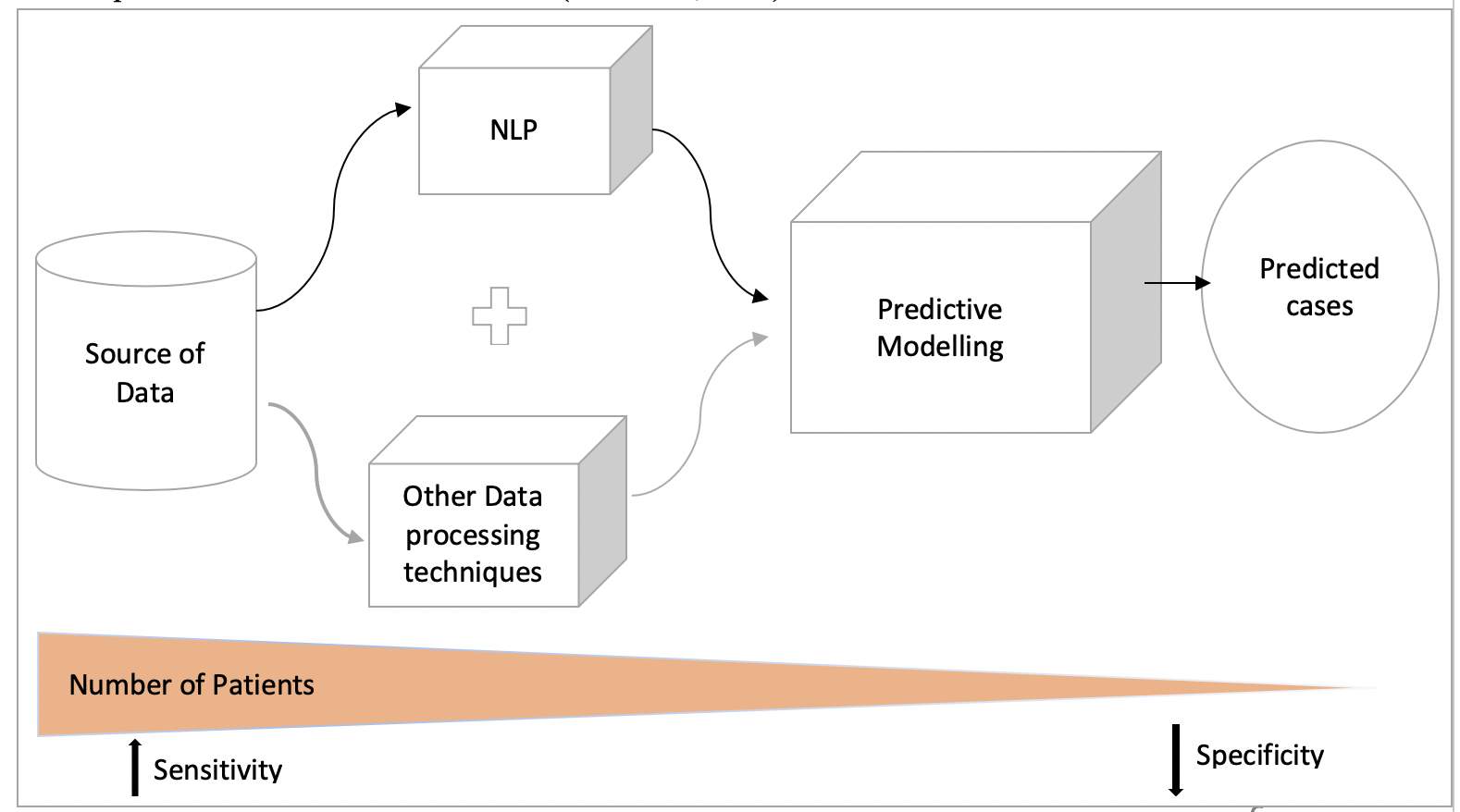
Figure 3: An overview of data processing . (Liao et al., 2015).
In the past few years, a number of studies for understanding the trend of specific disease in a given population, have leveraged the processing power of various NLP concepts (Imler, Morea, & Imperiale, 2014). Like any other concept, NLP has various concepts that can be used strategically for a specific task. In order to process the free text available in medical record, one of the most used NLP concepts is Bag of words (bow) model. In order to predict the cases of sepsis in an emergency department, bow model was used to tokenize and transform the data. The data was then processed using a predictive model and the cases of sepsis were accurately recognized (Horng et al., 2017). An automated trigger was then designed for the emergency department using the same NLP and machine learning concepts. This study demonstrated the use of bow model in a predictive algorithm with clinical notes and showed promising result (Long, 2018). In order to determine the predictive value of the clinical notes, the bag of word model was used to extract information and successfully understand the representation of words in a clinical context (Boag, n.d.).
Another NLP model used to process clinical notes is topic modeling. Topic modeling was used to train clinical documents as a query expansion method and showed best performance for retrieving clinically relevant documents (Zeng, Redd, Rindflesch, & Nebeker, 2012). The F1- scores of topic model was better than the other two models used for term expansion. Topic modeling also showed promising results on an investigative analysis done using narrative notes. Topic modeling used with a Bayesian predictive model accurately predicted cases of high risk patients in an intensive care unit (Luo & Rumshisky, 2017). Latent Dirichlet allocation (LDA), a type of topic modeling approach was used in predicting medication and diagnosis in the National Taiwan University Hospital, Taiwan (Lu, Wei, & Hsiao, 2016) . The model outperformed logistic regression and k-nearest-neighbor prediction model while processing the healthcare data.
Word embeddings is another NLP model that is used in clinical natural language processing. A qualitative and quantitative analysis done on clinical notes demonstrated the ability of word embeddings in vector representation and capturing linguistic relationships between words (Wang et al., 2018). The dataset trained by word embeddings showed more accurate clinical results than the ones from Google news and GloVe (Wang et al., 2018). Another research study to understand the mortality rate in an Intensive Care unit used word2vec, a word embeddings model, to be incorporated in the predictive neural network method (Grnarova, Schmidt, Hyland, & Eickhoff, 2016). The research concluded that understanding linguistic relations of the words in the caregivers notes is a vital part of predictive models and needs to be accounted to reduce patient mortality (Grnarova et al., 2016) .
METHOD
For the following project, natural language processing concepts discovered by the help of literature review will be used to run some preliminary analysis on healthcare data and propose what can be the future steps to improve the specificity and sensitivity of CDS towards sepsis and reduce alerts fatigue. We will load the data in Jupyter, which is an online, open source software that allows the use of different coding languages and we will use Python to process NLP on the data (“Project Jupyter,” n.d.). Our focus would be to explore the NLP concept using caregivers notes and then suggest future supervised or unsupervised predictive model that would improve the CDS.
Text parsing and exploratory analysis
Modelling
Text pre-processing
Text representation
Data Collection




Figure 4: A general representation of the steps involved. The boxes in orange represent the steps involved in NLP.
As per the literature review done, three NLP concepts identified to be effective in processing unstructured data from physician and RN notes were Bag of Words, Word embeddings, Topic modeling. The models are individually defined below:
- Bag of Words:
The bag of words (bow) model of NLP is used for representing the words/text by object categorization. Object categorization is the method of training a classifier to recognize a certain image and determine its category (Pinz, 2005). When used with python, in simpler words, the bag of word model extracts the frequency of a word in a string (Long, 2018).The way bow understands and categorizes the object is by a series of step. The first step in the model is to define the vocabulary and arranging them as vectors. The next step is to convert the vector into frequency vectors. The result is a bow model with frequency of each word in the model. For example, the inserted strings in the bow algorithm are: String 1 – “Discharge from ER”. String 2 – “Diagnosis and Discharge”.
Output: For string 1 = Discharge 1
Diagnosis 0
From 1
ER 1
And 0

Frequency of words
Convert to vectors
Design vocabulary
Collect data



Figure 5: Steps involved in a Bag of word model
- Topic Model:
Topic modeling is a type of NLP where the model believes that the relationship between the words depends on the category of the topic it falls into. Latent Dirichlet allocation (LDA) is a type of topic modeling algorithms which assumes that different documents represent different topics and the words are generated based on these topics (Chan et al., 2013). It uses a neural network approach of tracing back the words association with the topic (Chan et al., 2013). Topic model also uses the bag of word approach to then finally understand the frequency of words in the topics discovered.
Clustering of words

Topic model
Collect Data
Distribution of topics across the data



Frequency of words in the topics
Figure 6: Steps involved in a simple topic model algorithm
- Word Embeddings:
Word embedding is another kind of NLP model, that is used for representing the vocabulary by understanding the context of a word in a document (Wang et al., 2018). Similar to bow, Word embeddings is used to vectorize the words and place the words in a spatial dimension, closer to each other depending on the similarity of the context (Wang et al., 2018). The concept of this method is very similar to the bow model but with more emphasis on context of the words (Boag, n.d.).

Context Output
Word embedding algorithm
Collect Data


Figure 7: A simple demonstration of word embedding model.
RESULTS AND DISCUSSION:
For exploratory purposes, Bag of word model on extracted data using Python language. The following section would demonstrate how the model represented and processed the words in the caregiver’s notes.
System Specifications:
The NLP concept was run using Python 3 language on Jupyter Notebook, an online open source application.
Data Collection:
The caregiver’s notes data was downloaded from the MIMIC-III database. This dataset is ideal for our exploratory research as it is the de-identified data of patients in ICU, which is where the highest prevalence for sepsis, severe sepsis and septic shock is found (Mao et al., 2018).For recognizing Sepsis in a patient, doctors diagnose a number of physical findings such as Low blood pressure, increased heart rate, difficulty breathing, and some lab tests that check for organ damage or visible signs of infection. We will use a combination of care-givers notes, lab data and procedures data from the data set. The bag of words python code was reproduced by following a few examplesTo determine the Gold standards for Sepsis the following criteria was considered. (Calvart et al., 2016):
- Presence of ICD-9 codes related to an infection.
- Systolic blood pressure < 90 mmHg for at least 60 minutes.
- Total fluid replacement >= 1200mL or >= 20mL/kg for 24 hours.
- SIRS criteria score >=2.
Bag of Word model:
| Functions | Libraries used |
| Data manipulation | Pandas |
| Array formation and processing | Numpy |
| Tokenization and vectorization of words | nltk |
| For regular expressions | re |
Table 1: Python Libraries used for BOW model
- Text Preprocessing: Loading and understanding the data:
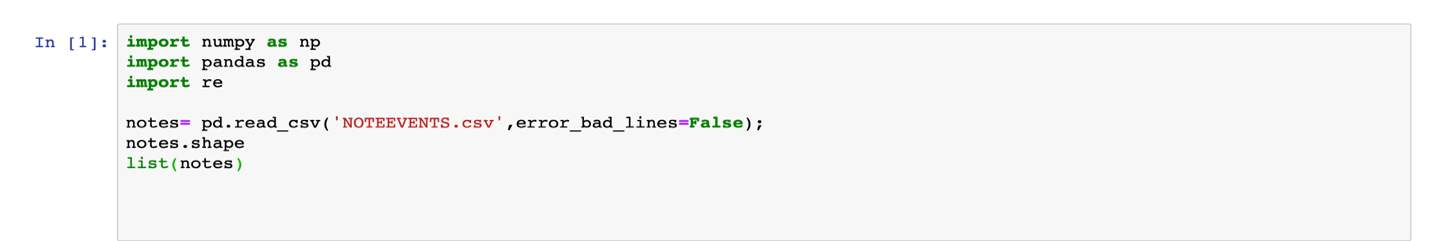
Figure 8: Python code for loading the document and viewing the column headers of the file.

Figure 9: Output- The columns present in the ‘Noteevents.csv’ file as opened in Jupyter Notebook.
Figure 10: Loading the ‘LABEVENTS.csv” -Lab records and viewing the column

Figure 11: Loading Procedures dataset and checking for specific ICD9 codes associated with Sepsis. For the exploratory purposes’ patient data with codes 0202,3995 and 458 were selected.

Figure 12: After loading the datasets and merging them the data was randomly divided into validation set, training set and test set to be further used for the predictive model.

Figure 13: The data sets were preprocessed to remove spaces, replace newlines and carriage return to fit the data into a perfect data frame and for easier tokenization of the data.
Figure 14: Vectorization of the processed data.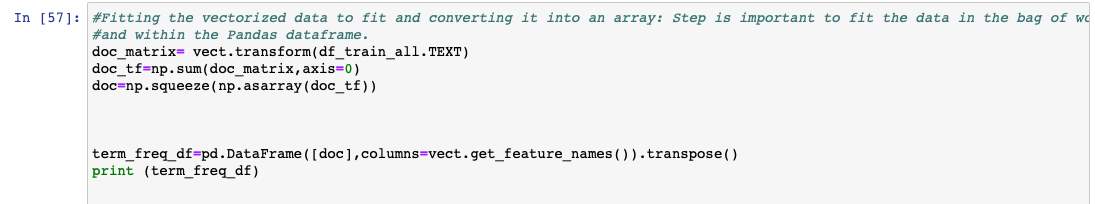
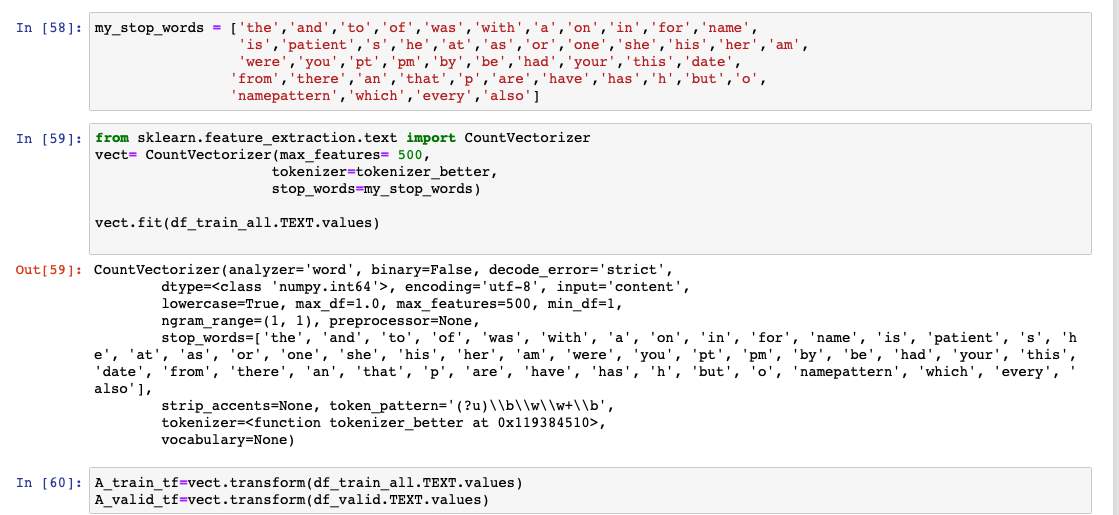

Figure 14: After fitting the data for Vectorizing and transforming into array if we print one of the sets, we get the frequency/count of the words. This data is then used to build a predictive model.
Future steps:
After the step of processing the data, a standard Machine learning technique could be used to weigh the results of the models used. Logistic regression could be the simplest approach taken for building a predictive model (Long, 2018).Research studies based on building predictive models for identifying patients at risk of sepsis infection concluded that a Bag of Word model of free text, triage data and vital signs data outperformed in improving specificity and sensitivity of the model (Fig) (Horng et al., 2017). 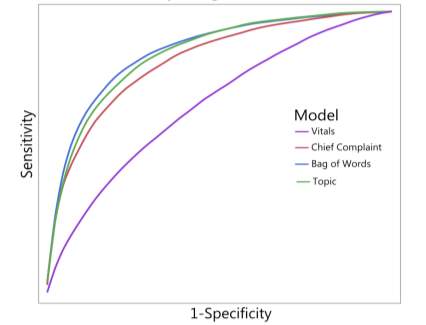
Figure : An receiver operating characteristics curve showing Bow model and topic model outperforming other models with just structured data (Horng et al., 2017).
Similarly, a predictive model build using Bag of words (NLP model) and Support Vector Machine (supervised machine learning), analyzed on two different datasets concludes that the specificity and sensitivity of the system increases as the entire dataset is taken into consideration (Weng, Wagholikar, McCray, Szolovits, & Chueh, 2017). Together with machine learning and data mining techniques, a system can be made specific and sensitive for a required task.
CONCLUSION:
According to Coleman et.al sensitivity of a system is described by the ability of the CDS to alert caregivers when patients are at risk and specificity of the CDS system is its ability to give less false positives (Coleman et al., 2013). Implementing a knowledge based, high specific algorithm can possibly bridge the gap. Most of the CDS system still focuses on the structured data, neglecting the unstructured physician notes that is said to contain time sensitive information. NLP can change the game with its focus on representation of words and context-based analysis of the unstructured data. The data processing done by NLP standardizes the unstructured data by breaking the dataset into words, vectorizing them, and then finding the frequency of the words in the dataset. The words are then divided into different training and test sets to be used in a predictive model. Using this predictive model, we predict specific disease prevalence or at risk patient demographics. As the knowledge base build for CDS leverages the information stored in the caregiver’s notes using deep learning techniques, it makes the system more sensitive and specific to the date and hence sends out alerts accordingly. Not only does this predict accurate cases for efficient care delivery but also doesn’t give out unnecessary alerts.
ACKNOWLEDGEMENT:
REFERENCE:
Angus, D. C., Linde-Zwirble, W. T., Lidicker, J., Clermont, G., Carcillo, J., & Pinsky, M. R. (2001). Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Critical Care Medicine, 29(7), 1303–1310.
Apostolova, E., & Velez, T. (2018). Toward Automated Early Sepsis Alerting: Identifying Infection Patients from Nursing Notes. ArXiv:1809.03995 [Cs]. Retrieved from http://arxiv.org/abs/1809.03995
Bates, D. W., Kuperman, G. J., Wang, S., Gandhi, T., Kittler, A., Volk, L., … Middleton, B. (2003). Ten Commandments for Effective Clinical Decision Support: Making the Practice of Evidence-based Medicine a Reality. Journal of the American Medical Informatics Association, 10(6), 523–530. https://doi.org/10.1197/jamia.M1370
Boag, W. (n.d.). What’s in a Note? Unpacking Predictive Value in Clinical Note Representations, 9.
Carli, D., Fahrni, G., Bonnabry, P., & Lovis, C. (2018). Quality of Decision Support in Computerized Provider Order Entry: Systematic Literature Review. JMIR Medical Informatics, 6(1). https://doi.org/10.2196/medinform.7170
Carrell, D. S., Halgrim, S., Tran, D.-T., Buist, D. S. M., Chubak, J., Chapman, W. W., & Savova, G. (2014). Using Natural Language Processing to Improve Efficiency of Manual Chart Abstraction in Research: The Case of Breast Cancer Recurrence. American Journal of Epidemiology, 179(6), 749–758. https://doi.org/10.1093/aje/kwt441
Chan, K. R., Lou, X., Karaletsos, T., Crosbie, C., Gardos, S., Artz, D., & Rätsch, G. (2013). An Empirical Analysis of Topic Modeling for Mining Cancer Clinical Notes. In 2013 IEEE 13th International Conference on Data Mining Workshops (pp. 56–63). https://doi.org/10.1109/ICDMW.2013.91
Coleman, J. J., van der Sijs, H., Haefeli, W. E., Slight, S. P., McDowell, S. E., Seidling, H. M., … Slee, A. (2013). On the alert: future priorities for alerts in clinical decision support for computerized physician order entry identified from a European workshop. BMC Medical Informatics and Decision Making, 13, 111. https://doi.org/10.1186/1472-6947-13-111
Collins, S. (2018). Project Information – NIH RePORTER – NIH Research Portfolio Online Reporting Tools Expenditures and Results. Retrieved October 12, 2018, from https://projectreporter.nih.gov/project_info_description.cfm?aid=9678555&icde=39574855&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC&pball
Dale, J., Barbee James, M., Casey, P., & Stein, B. (2016). Integrated Clinical Decision Support Focused on Early Recognition and Standardized Treatment of Sepsis. Retrieved October 21, 2018, from https://www.shmabstracts.com/abstract/integrated-clinical-decision-support-focused-on-early-recognition-and-standardized-treatment-of-sepsis/
Demner-Fushman, D., Chapman, W. W., & McDonald, C. J. (2009). What can natural language processing do for clinical decision support? Journal of Biomedical Informatics, 42(5), 760–772. https://doi.org/10.1016/j.jbi.2009.08.007
Grnarova, P., Schmidt, F., Hyland, S. L., & Eickhoff, C. (2016). Neural Document Embeddings for Intensive Care Patient Mortality Prediction. ArXiv:1612.00467 [Cs]. Retrieved from http://arxiv.org/abs/1612.00467
Horng, S., Sontag, D. A., Halpern, Y., Jernite, Y., Shapiro, N. I., & Nathanson, L. A. (2017). Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLOS ONE, 12(4), e0174708. https://doi.org/10.1371/journal.pone.0174708
Imler, T. D., Morea, J., & Imperiale, T. F. (2014). Clinical decision support with natural language processing facilitates determination of colonoscopy surveillance intervals. Clinical Gastroenterology and Hepatology: The Official Clinical Practice Journal of the American Gastroenterological Association, 12(7), 1130–1136. https://doi.org/10.1016/j.cgh.2013.11.025
Joshi, A. K. (1991). Natural Language Processing. Science, 253(5025), 1242–1249. https://doi.org/10.1126/science.253.5025.1242
Kesselheim, A. S., Cresswell, K., Phansalkar, S., Bates, D. W., & Sheikh, A. (2011). Clinical Decision Support Systems Could Be Modified To Reduce ‘Alert Fatigue’ While Still Minimizing The Risk Of Litigation. Health Affairs, 30(12), 2310–2317. https://doi.org/10.1377/hlthaff.2010.1111
Ledley, R. S., & Lusted, L. B. (1959). Reasoning Foundations of Medical Diagnosis. Science, New Series, 130(3366), 9–21.
Liao, K. P., Cai, T., Savova, G. K., Murphy, S. N., Karlson, E. W., Ananthakrishnan, A. N., … Kohane, I. (2015). Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ, 350, h1885. https://doi.org/10.1136/bmj.h1885
Long, A. (2018, June 4). Introduction to Clinical Natural Language Processing: Predicting Hospital Readmission with…. Retrieved November 15, 2018, from https://towardsdatascience.com/introduction-to-clinical-natural-language-processing-predicting-hospital-readmission-with-1736d52bc709
Lu, H.-M., Wei, C.-P., & Hsiao, F.-Y. (2016). Modeling healthcare data using multiple-channel latent Dirichlet allocation. Journal of Biomedical Informatics, 60, 210–223. https://doi.org/10.1016/j.jbi.2016.02.003
Luo, Y.-F., & Rumshisky, A. (2017). Interpretable Topic Features for Post-ICU Mortality Prediction. AMIA Annual Symposium Proceedings, 2016, 827–836.
Mao, Q., Jay, M., Hoffman, J. L., Calvert, J., Barton, C., Shimabukuro, D., … Das, R. (2018). Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open, 8(1). https://doi.org/10.1136/bmjopen-2017-017833
Marik, P. E., & Taeb, A. M. (2017). SIRS, qSOFA and new sepsis definition. Journal of Thoracic Disease, 9(4), 943–945. https://doi.org/10.21037/jtd.2017.03.125
McCoy, A. B., Thomas, E. J., Krousel-Wood, M., & Sittig, D. F. (2014). Clinical Decision Support Alert Appropriateness: A Review and Proposal for Improvement. The Ochsner Journal, 14(2), 195–202.
Ohno-Machado, L. (2011). Realizing the full potential of electronic health records: the role of natural language processing. Journal of the American Medical Informatics Association: JAMIA, 18(5), 539. https://doi.org/10.1136/amiajnl-2011-000501
Pinz, A. (2005). Object Categorization. Foundations and Trends® in Computer Graphics and Vision, 1(4), 255–353. https://doi.org/10.1561/0600000003
Project Jupyter. (n.d.). Retrieved December 4, 2018, from https://www.jupyter.org
Sarkar, D. (DJ). (2018, June 19). A Practitioner’s Guide to Natural Language Processing (Part I) — Processing & Understanding Text. Retrieved November 15, 2018, from https://towardsdatascience.com/a-practitioners-guide-to-natural-language-processing-part-i-processing-understanding-text-9f4abfd13e72
Savova, G. K., Fan, J., Ye, Z., Murphy, S. P., Zheng, J., Chute, C. G., & Kullo, I. J. (2010). Discovering Peripheral Arterial Disease Cases from Radiology Notes Using Natural Language Processing. AMIA Annual Symposium Proceedings, 2010, 722–726.
Savova, G. K., Masanz, J. J., Ogren, P. V., Zheng, J., Sohn, S., Kipper-Schuler, K. C., & Chute, C. G. (2010). Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association, 17(5), 507–513. https://doi.org/10.1136/jamia.2009.001560
Sittig, D. F., Ash, J. S., & Ledley, R. S. (2006). The Story Behind the Development of the First Whole-body Computerized Tomography Scanner as Told by Robert S. Ledley. Journal of the American Medical Informatics Association : JAMIA, 13(5), 465–469. https://doi.org/10.1197/jamia.M2127
Sittig, D. F., Wright, A., Osheroff, J. A., Middleton, B., Teich, J. M., Ash, J. S., … Bates, D. W. (2008). Grand challenges in clinical decision support. Journal of Biomedical Informatics, 41(2), 387–392. https://doi.org/10.1016/j.jbi.2007.09.003
Wang, Y., Liu, S., Afzal, N., Rastegar-Mojarad, M., Wang, L., Shen, F., … Liu, H. (2018). A Comparison of Word Embeddings for the Biomedical Natural Language Processing. Journal of Biomedical Informatics. https://doi.org/10.1016/j.jbi.2018.09.008
Weng, W.-H., Wagholikar, K. B., McCray, A. T., Szolovits, P., & Chueh, H. C. (2017). Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Medical Informatics and Decision Making, 17. https://doi.org/10.1186/s12911-017-0556-8
Wright, A., & Sittig, D. F. (2008). A four-phase model of the evolution of clinical decision support architectures. International Journal of Medical Informatics, 77(10), 641–649. https://doi.org/10.1016/j.ijmedinf.2008.01.004
Zeng, Q. T., Redd, D., Rindflesch, T., & Nebeker, J. (2012). Synonym, Topic Model and Predicate-Based Query Expansion for Retrieving Clinical Documents. AMIA Annual Symposium Proceedings, 2012, 1050–1059.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Health"
Health is the general condition of the body or mind. The World Health Organization defines health as “a state of complete physical, mental and social well-being and not merely the absence of disease or infirmity.”
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




