Recombinant Mussel Adhesive Proteins (rMAPs)
Info: 6530 words (26 pages) Dissertation
Published: 9th Dec 2019
Tagged: BiologyMarine Studies
Table of Contents
2. Problem Statement and Hypothesis
4.1. Mussels Adhesive Mechanism
4.2. Natural Extraction of Mussel Adhesive Proteins (MAPs)
4.3. Recombinant Mussel Adhesive Proteins (rMAPs)
4.3.1. Overview of Recombinant Protein Expression
4.3.3. Protein Fusion Approach
4.3.4. Modification of rMAPs with Tyrosinase
4.4. Further Enhancement of rMAP Adhesive Property
4.4.2. Multi-protein Nanofibres
4.5. Application of adhesive from Mussel protein
The ability of mussel to stick strongly to surfaces underwater has attracted a lot of attention, especially in the engineering world. It has been found that mussel produce a protein which act as a glue. The mussel adhesive proteins or mussel foot proteins (mfps) contain amino acid 3,4-dihydroxyphenylalanine (DOPA) that play a key role in the adhesive strength of mussel.
Initially, mfps were obtained by naturally extracting the proteins from mussels. However, this method is labour intensive and very inefficient, requiring an estimated 10,000 mussels to extract up to 1g of mfp (Cha et al., 2008). The low extraction yield and high production cost of natural extraction restricts the application of MAPs to small scale applications. Recombinant DNA technology was applied to overcome this problem. Recombinant mussel foot proteins were successfully produced from the gene sequences of mfps that were isolated from mussels (Hwang et al., 2005, Hwang et al., 2004). This indicates the potential for mass production of mfps to increase practical applications as an adhesive.
Post-translational modification using tyrosinase enzyme becomes essential as the mfps produced via prokaryotic host does not undergo such process. The recombinant mfp lacks some properties like the one extracted naturally. The product functionality does not possess the same quality as intended
Some drawbacks of recombinant mfps suchs as low production and purification yields have prompted research on protein fusion approaches to overcome said drawbacks (Hwang et al., 2004). One of the fusion approaches involves fusing an mfp with a different mfp, for example fusing mfp-1 with mfp-5 proteins to form a hybrid mfp-151 (Hwang et al., 2007a). Another fusion method builds on the former approach, by fusing the hybrid protein with a peptide to form a protein-peptide hybrid (Hwang et al., 2007b). This protein-peptide hybrid was found to not only retain the advantages of the hybrid protein, but also shows superior adhesive ability. Thus, fusing mfps with peptides appears promising in improving recombinant mfp adhesiveness.
Producing a fusion of mfps and protein nanofibers have also shown a promising result. The amyloid-based protein displays unique properties such that when combined with dopa-based protein, the two functionality is reflected in the new fusion protein. Importantly, the strength of adhesiveness is at the unprecedented level, about 1.5 times higher than any bio-inspired protein.
2.1. Problem Statement
Currently, the adhesiveness of recombinant MAPs (rMAPs) is still not as strong as naturally extracted MAPs. Reasons for the low adhesiveness of rMAPs could be attributed to the modification of tyrosine residues into DOPA via tyrosinase enzyme, and the low cohesion due to weak cross-linking during modification. Existing enzymatic modification methods using the tyrosinase have low modification yield and the modified MAPs have not yet delivered the same quality as those obtained from natural extraction (Jeon et al., 2015).
2.2. Hypothesis
The adhesiveness of recombinant MAPs can be improved by using a protein fusion approach with peptides.
3.1. Project Objectives
The key objectives include the expression of recombinant mussel foot proteins (mfps) and enzyme tyrosinase, enzymatic modification of the recombinant mfps using the tyrosinase expressed, and conducting tests on the adhesive functionality of the modified products.
3.2. Research Scope
This project focuses on improving the adhesive properties of recombinant mfps by fusion with peptides. Specifically, recombinant mfp-5 and mfp-3 will be expressed and fused to R3 and T26 peptides respectively. Bacteria Escherichia coli (E. coli) is used for expression system. Currently, treating with tyrosinase enzyme for post-translational modification of the rMAPs poses drawbacks such as low modification yield as well as weak crosslinking. The effect of fusing proteins with peptides will be investigated by conducting tests on the recombinant proteins before and after modifying with tyrosinase. The functionality of the two fusion products should be compared in similar environments.
4.1. Mussels Adhesive Mechanism
Mussels can stick to surfaces underwater due to mussel foot protein (mfp) as produced by the mussel byssus (Castillo et al., 2017). Mussel byssus can be classified into four parts: root, stem, threads, and attachment plaques (Castillo et al., 2017). Attachment plaque has an essential role to give rise to adhesiveness (Castillo et al., 2017).
Brown mussel (Mytilus edulis) is the most popular choice of species for mussel adhesive proteins extraction (Castillo et al., 2017). The characteristics of mussel adhesive proteins are easy to be digested in nature and capable of wet adhesion (Castillo et al., 2017). There are six different mussel foot proteins that have a unique characteristics mainly mfp-1, mfp-2, mfp-3, mfp-4, mfp-5, and mfp-6. Among them, mfp-5 has the most abundant DOPA concentration (Castillo et al., 2017).
Protein adhesion energy has a direct relationship with pH (Wilker, 2011). MAPs are functional at acidic environment as DOPA is instantly oxidized to dopaquinone in neutral or alkali environment (Castillo et al., 2017).
The catechol side chain of the DOPA (ie. 1,2-dihydroxybenzene) can form bond with surfaces through hydrogen bonding and metal chelation (Wilker, 2015). The cation-cathecol combination in the mussel adhesive proteins have a significant role in the mussel adhesive mechanism, as compared to only catechols and only cations (Wilker, 2015). The adhesion energies in saline condition were at utmost level when catechol and cations are bundled together (Wilker, 2015).
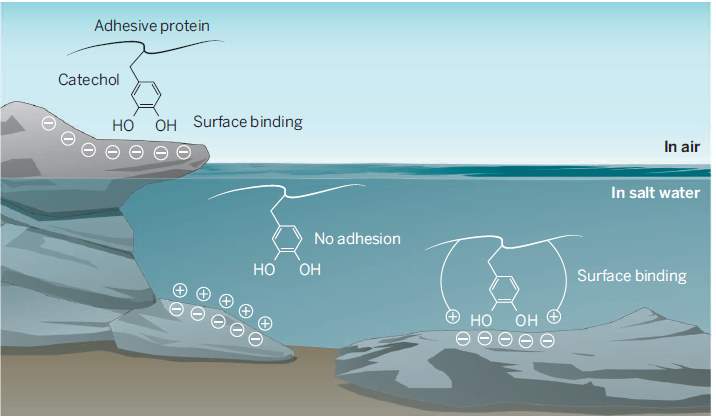
Figure 1 Catechol-cation mechanism (Wilker, 2015).
4.2. Natural Extraction of Mussel Adhesive Proteins (MAPs)
Two methods of obtaining mfps that are discussed in this report are natural extraction and recombinant production. Natural extraction of mfp involves the isolation of mfps directly from mussels collected and kept at regulated pH levels and low temperatures (Castillo et al., 2017). Mfps can be extracted from the plaques or from the feet of mussel byssus. Mussel plaques are secreted by the collected mussels when allowed to attach to a surface. The secretions are allowed to accumulate for up to 24 hours before being removed and collected (Papov et al., 1995). Secretion of mussel plaques can also be induced by introducing potassium chloride into the mussel feet (Gantayet et al., 2013, Tamarin et al., 1976). Collection of mussel feet, on the other hand, is accomplished by directly cutting the feet off fresh mussels that have been drained of blood and immediately frozen (Papov et al., 1995).
Mfps can then be selectively isolated from the collected feet and plaques through a series of steps (Castillo et al., 2017). Hwang et al. obtained mfps using a process consisting of three steps (Hwang et al., 2010). These steps are similar to the steps involved in cell fractionation and include homogenisation to form an emulsion, centrifugation of the homogenates to separate out the mfps which are collected. The denser remnants that settled out during centrifugation are subjected to further homogenisation and centrifugation until all mfps have been extracted (Steele, n.d., Hwang et al., 2010, Castillo et al., 2017). Once mfps have been isolated from the source mussels, purification is required to ensure high purity of naturally extracted mfp. Purification is carried out chromatographically via methods such as liquid, gel filtration, size exclusion, ion-exchange and affinity chromatography (Waite, 1995, Ohkawa et al., 2009, Xu et al., 2014, Castillo et al., 2017).
A significant obstacle in the natural extraction of MAPs is its extremely low efficiency coupled with high labour demand. An estimated 10,000 mussels were required to obtain a mere 1 g of adhesive protein (Cha et al., 2008, Castillo et al., 2017). The low mfp content in mussels, low efficiency of mfp extraction techniqes and low purification yield collectively contribute to the low extraction yield of mfps from mussels (Castillo et al., 2017).
4.3. Recombinant Mussel Adhesive Proteins (rMAPs)
The low extraction yield and costly production of mfps obtained via natural extraction are detrimental to the practical application of MAPs, restricting use to small scale applications (Cha et al., 2008). In order to overcome this limitation, recombinant DNA technology was adapted to artificially mass-produce MAPs.
4.3.1. Overview of Recombinant Protein Expression
Protein expression refers to the process of producing of a protein from a gene (Scientific, n.d.-b). The foundation of recombinant protein expression is defined by the central dogma of biology, which describes the transmission of genetic information from DNA, to RNA, to construct a protein. DNA is a molecule consisting of two chains in a double helix structure, and carries information required to produce proteins. RNA is similar to DNA with the exception of having a single helix structure, and functions as a messenger that delivers the information transcribed from DNA to be translated into a specific protein (Scientific, n.d.-a, Rettner, 2017).
The key steps of recombinant protein expression are cloning the gene sequence of interest into an expression vector, transformation of the expression host, selection of the transformed hosts and production of protein. First of all, the gene sequence of the protein to be cloned and the expression vector are isolated. A suitable vector should possess a combination of a selection marker, strong promoters, ribosome binding sites, termination sequence, affinity tag and multi-enzyme restriction site (Scientific, n.d.-b).The function of a selection marker is to grant the expression host resistance to antibiotics, the promoters enhance the expression of the gene of interest, ribosome binding sites are essential for the translation of messenger RNA, termination sequences allow for control transcription and affinity tags assist in protein purification (Scientific, n.d.-c, Clark and Pazdernik, 2016, Hwang et al., 2004). The isolated gene is inserted into the vector, which is then introduced into an expression host by transformation. Subsequently, the transformed hosts undergo cloning in a medium that contains antibiotics. Only the hosts that possess the antibiotics resistance selection marker will grow, and recombinant proteins can be induced and produced (Griffiths et al., 2000).
4.3.2. Expressing rMAPs
In expression of recombinant mussel adhesive proteins (rMAPs), the gene sequence of interest is isolated from mussels inserted into an expression vector, typically a bacterial plasmid. Over the years, protein expression with various expression hosts were attempted using both prokaryotic (nucleus-lacking) and eukaryotic (nucleus-containing) species. An early effort of expressing mfps utilised eukaryotic yeast cells, specifically a D8 yeast strain, to produce mefp-1 (mfp-1 extracted from a Mytilus edulis mussel) with a maximum yield of 5% of total proteins produced (Filpula et al., 1990). Insect cells have also been experimented with, where functional rMAPs were produced in insect Sf9 cells (Lim et al., 2011).
As of today, Escherichia coli (E. coli) is the most commonly used prokaryotic expression host in rMAP expression with the usage of E. coli BL21 (DE3) host strain being a common theme (Hwang et al., 2005, Wong et al., 1998, Kitamura et al., 1999, Zeng et al., 2010, Yang et al., 2013, Nicklisch et al., 2013, Hwang et al., 2007a). Studies have reported successful expression of recombinant mefp-1, mgfp-3, mgfp-5 and mcfp-6 proteins with functional adhesive properties in E. coli expression systems (Zeng et al., 2010, Hwang et al., 2005, Hwang et al., 2004, Nicklisch et al., 2013). Recombinant mgfp-5 proteins in particular were reported to exhibit strong adhesive ability, surpassing that of Cell-Tak, a commercialised MAP-based product comprising of natural extraction of MAPs (Hwang et al., 2004, Cha et al., 2008).
However there were a number of drawbacks to the recombinant mgfp-5, for example the expression of the rMAP in soluble form hindered cell growth and led to a low production yield (Cha et al., 2008, Hwang et al., 2007a). Purification yield of the recombinant protein was also low as its inherently adhesive nature caused it to adsorb (strongly attach) to the resin used in the purification process (Cha et al., 2008, Hwang et al., 2007a, Hwang et al., 2004). The recombinant mgfp-5 was also found to be very insoluble after being purified, thus developing a concentrated solution of mgfp-5 for practical applications was unachievable (Cha et al., 2008, Hwang et al., 2007a). Additionally, degradation of expressed mgfp-5 was observed, indicating that mgfp-5 is toxic to E. coli (Hwang et al., 2004). In order to overcome these problems, efforts have been made to explore hybrid protein fusion approaches.
4.3.3. Protein Fusion Approach
Due to the limitations of recombinant mfp-5 expression, a recombinant hybrid fusion of mfp-1 and mfp-5 proteins was developed based on mfp-5 because it has demonstrated high adhesiveness in comparison with commercial Cell-Tak (Hwang et al., 2004). The mfp-151 hybrid exhibited significantly improved production yield of ~40% of total proteins in comparison with mfp-5 (~13%) (Hwang et al., 2007a). The purification yield of mfp-151 was also much greater than that of mfp-5, with a final purification yield of ~53% compared to ~7% (Cha et al., 2008, Hwang et al., 2004). The recombinant hybrid protein showed high post-purification solubility, where mfp-5 was highly insoluble, allowing mfp-151 to be concentrated into a sufficiently viscous solution (Hwang et al., 2007a). This suggests a possibility of bulk-scale economical production of rMAP-based adhesives for practical usage. The expression of mfp-151 as insoluble inclusion bodies further overcomes the limitations of mfp-5 as it it no longer toxic to E. coli cells and can be expressed in large quantities (Hwang et al., 2007a). In addition to improved adhesive abilities, the recombinant hybrid mfp-151 also showed high biocompatibility for various cell types, adding to its potential as a bioadhesive for cells or tissues (Hwang et al., 2007b, Hwang et al., 2007a, Hwang et al., 2004).
4.3.4. Modification of rMAPs with Tyrosinase
Tyrosinases are copper-containing enzymes which are available in many living things (Halouli et al., 2005). The enzyme catalyses the ortho-hydroxylation of monophenols (monophenolase activity) and further the oxidation of o-diphenols to o-quinones (diphenolase activity) using molecular oxygen (Halouli et al., 2005). Tyrosinase is the cause of the early mechanism of melanin synthesis from L-tyrosine leading to the formation of L-dopaquinone and L-dopachrome (Halouli et al., 2005).
There are two different sources that Tyrosine can be derived from in this project which are Bacillum megaterium and Pycnoporus species. Also, there are two different methods to express Tyrosinase, using natural extraction as well as recombinant DNA.
In natural extraction, the source is grown in a medium that contains nutrient-growth materials. The tyrosinase enzyme will be produced along with the growth in number of strain. Cell lysates were obtained and cell debris were removed via centrifugation to obtain the supernatant for enzymatic activity assay (Halouli et al., 2005). This experiment was done using Pycnoporus strain.
As elaborated earlier on how to create recombinant DNA on section 4.3, tyrosinase gene is obtained and selected just like rMAPs. Primers which included the NcoI and BglII restriction sites and a His6-tag in the C-terminus were designed and used to amplify the gene from the genomic DNA. (Shuster and Fishman, 2009). Only the batch that has Tyrosinase gene is harvested.
Although MAPs are expressed using optimized system and expression conditions, the adhesiveness was much lower than one via natural extraction (Castillo et al., 2017). When expressed in prokaryote host, only tyrosine residue contained in rMAPs and it requires additional modification (Castillo et al., 2017). In the role of mussel adhesiveness, tyrosinase is needed at the final step of post-translational modification if rMAPs is expressed in E. coli(Castillo et al., 2017). Tyrosine residues in mfps are converted to 3,4-dihydroxyphenylalanine (DOPA) (Castillo et al., 2017). This amino acid is important to produce mussel’s adhesive matrix of cross-linked proteins (Wilker, 2011). In the natural environment, mussels secrete a mixture of proteins that contains DOPA (Wilker, 2015). There are two forces at work: cohesion and adhesion (Figure 2). Surface adhesion arises from catechol side chain of DOPA, which form hydrogen bonding and metal chelation with the surface. Cohesion via covalent bonding occurs through oxidation of DOPA to a semiquinone or a quinone (Wilker, 2015).
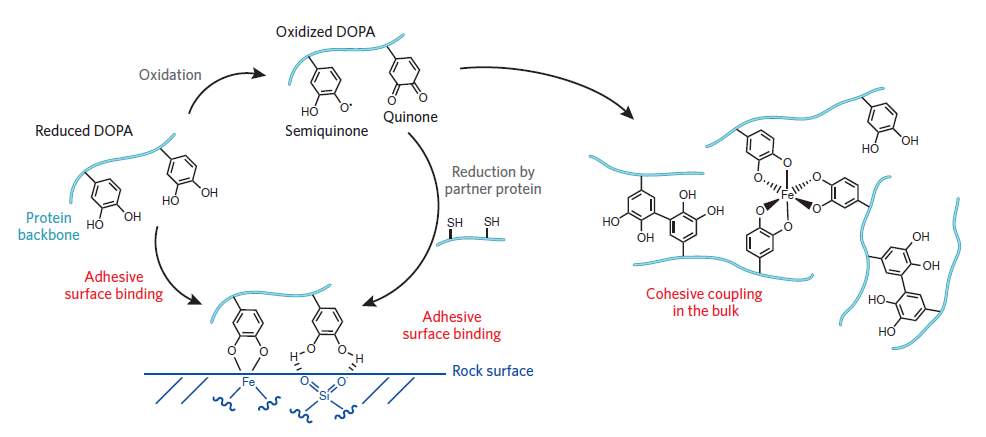
Figure 2 The cohesion and adhesion mechanism in mussel adhesive proteins (Wilker, 2011).
However, treating the rMAPs with tyrosinase for post-translational modification has not yet successfully delivered the expected result. There is a low modification yield and the quality of the products are still not on par with the mfp via natural extraction (Castillo et al., 2017).
4.4. Further Enhancement of rMAP Adhesive Property
4.4.1. Fusion with Peptides
In the efforts of hybrid fusion recombinant protein expression, it was established that the mfp-151 hybrid can be used as a bioadhesive for cells or tissues. A study was carried out with the objective of improving the cell adhesiveness of mfp-151 by fusion with an RGD peptide (Hwang et al., 2007b). This was achieved by adding a GRGDSP sequence to the C-terminus of the recombinant mfp-151 and expressing in an E. coli expression system to produce mfp-151-RGD insoluble inclusion bodies (Hwang et al., 2007b, Castillo et al., 2017).
It was found that both recombinant mfp-151 and mfp-151-RGD (mfp-RGD) had similar production yields and extraction yields, which implied that the mfp-RGD hybrid retained the advantages of the predecessor mfp-151 hybrid (Hwang et al., 2007b). The adhesive and spreading properties of mfp-RGF were investigated in comparison with mfp-151 and two commercial cell-adhesives, PLL and Cell-Tak (Hwang et al., 2007b). The mfp-RGD hybrid showed exceptional cell-adhesiveness and cell spreading, outranking all three other adhesive materials (Hwang et al., 2007b). It is suggested that the three binding mechanisms involved in the mfp-RGD hybrid is the reason for its superiority in cell-adhesion and cell spreading. These three binding mechanisms are described as the DOPA-mediated adhesion of Cell-Tak, the cationic cohesive interactions of PLL, and the RGD-sequence mediated fibronectin adhesion (Hwang et al., 2007b). Therefore, the recombinant hybrid mfp-151-RGD is can be successfully used as a cell adhesive in biotechnological applications such as in cell culture or tissue engineering.
4.4.2. Multi-protein Nanofibres
The method of generating rMAPs need to find a better solution or modification method. Other than dopa-based adhesives, some marine organisms shows ability for adhesion through amyloids. The effect of the combination of the two system is then tested for the integrated features from the two types of adhesive system as well as possibly imitate the intermolecular interactions between mfp3 and mfp5 observed in natural adhesion system (Zhong et al., 2014).
The combination of two independent natural adhesion systems which are DOPA-based adhesives and amyloid-based adhesive are tested (Zhong et al., 2014). Amyloids are β-strands that has fibrillar structures, they have intrinsic advantages for interfacial underwater adhesion (Zhong et al., 2014). Mfp-3 and Mfp-5 are selected from Mytilus galloprovincialis and CsgA are selected from Escherichia coli (Zhong et al., 2014). The fusion of proteins are CsgA-mfp3 and mfp5-CsgA that were engineered through isothermal one-step Gibson DNA assembly. CsgA-mfp3 and mfp5-CsgA would form into fibrous bundle due to CsgA being amyloidogenic (Zhong et al., 2014). The resulting fibres have ordered structures and multiple properties that are unique to both proteins. Interestingly, the strength of the adhesion energy in seawater achieve 20.9 mJ m-2., about 1.5 times greater than the capability of bio-engineered protein-based underwater adhesive so far (Zhong et al., 2014).
4.5. Application of adhesive from Mussel protein
Mussel adhesive proteins holds a tremendous potential for the development of the progress in biotechnology. Several applications involving mussel adhesive proteins have been developed such as to stifle soft-tissue inflammation (Service, 2017), as drug delivery for nanoparticle (News, 2018), and surface coating without physical chemical treatment (Weekly, 2016). The properties of mfp makes it a strong candidate for many biomedical application. Among some properties found are biocompatibility, pH-reactive, and ability to strongly bonded to metal ion (Weekly, 2016). The future application is vast and interesting for mussel adhesive protein especially for medical application (Weekly, 2016).
5.1. Approach and Methodology
For the expression of mfps, the recombinant E. coli is given with the mfp gene inserted. The E. coli was grown using either seed, auto-induction, or IPTG stock. Selection of good batch through introduction of antibiotic from YT media. Cell lysis is performed. Proteins need to be harvested by centrifuge and lysis. Lysis includes sonicate or lysis to break open. The bacteria that contains the gene-of-interest survives and thrives. Then, SDS-PAGE is be utilised to check for the mfp.
For the expression of tyrosinase enzyme,the recombinant E. coli with tyrosinase gene is inserted. Using either seed, auto-induction, or IPTG stock for growing E. coli. Selection of good batch through introduction of anti-biotic from YT media. Cell lysis is performed. Proteins need to be harvested by centrifuge and lysis. Lysis includes sonicate or lysis to break open. The bacteria that contains the gene-of-interest survives and thrives. Then, SDS-PAGE can be utilized to check for the tyrosinase protein.
Two options of purification will be carried out. The first option is to do column-free. There are two extraction method which are acetic-acid aided and urea-aided. In acetic-acid aided purification, after inclusion bodies are washed, acetic acid is added to obtain the MAP fusion proteins (Castillo, 2017). The mixture was left at 30 minutes at room temperatures then put in centrifuge and freeze-dried. The urea-aided used extraction buffer to resuspend IB, sonicate the mixture and left for 24 hours. The resulting mixture is a two-phase solution which consists of upper-clear part and gel-like part. The gel-like part is extracted and analyzed using SDS-PAGE. The second option is to do column-based via HiTrap. First, buffers and sample are prepared. The fractions are collected after each column equilibration and sample loading. Washing and eluting are required after each collection. SDS-page is performed to check the result and accuracy of the process in expressing mfp. Enzyme assay is used using 96 well plate and a reaction buffer to check for monophenolase and diphenolase activity.
To carry out enzyme modification, spectrophotometry is chosen to check the activity of enzyme on mfp. There will be a change in absorbance in relation to the modification yield as tyrosine is converted to dopa by enyzme tyrosinase.
Adhesive strength test will be performed using surface-coated or bulk adhesion. The surface coating analysis was used to observe the comparison between DOPA-rich mfp and DOPA-lacking mfp to stick to dry and wet surfaces. Some drops of modified and unmodified fusion protein are placed in dry and wet glass slide. All the slides are left to dry overnight and then washed with distilled water. The Coomassie blue staining solution are used to soak the glass slides to check for the presence of protein (Castillo, 2017). Bulk adhesion was executed through a shear strength test and consists of two parts. The first and second part of the test is done using a rough wood and a smooth glass surface respectively.
5.2. Project Plan
The proposed timeline of this project is presented in a Gantt chart as shown in Figure 3.
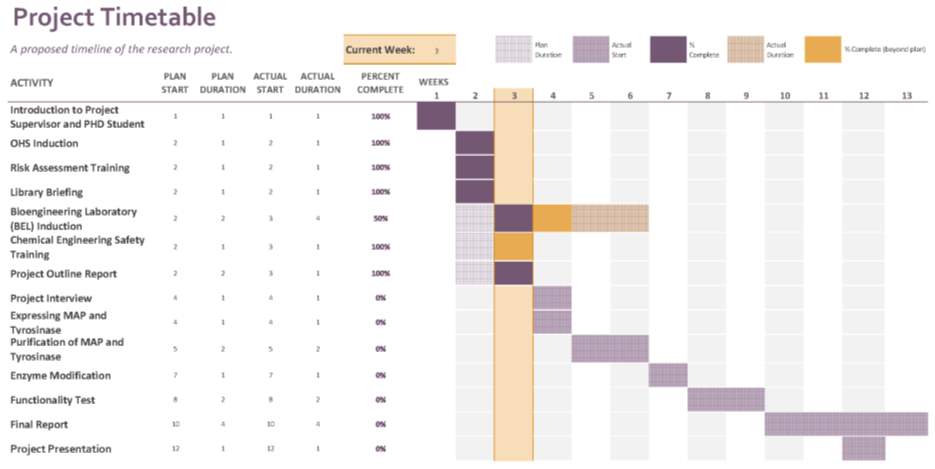
Safety needs to be upheld highly as some chemicals that are used in the experiment are hazardous. One of them is Coomassie blue staining solution. Nevertheless, the amount of chemicals that are considered hazardous are only a few compared to number of chemicals used in the experiment. Mostly, the likelihood of the hazard falls in unlikely. However, for those chemicals that possess some risk, controls are being put in place to ensure safe working condition. The complete risk assessment can be seen in appendix.
In terms of handling the equipment, most of the equipments have minor consequences. Only Autoclaves possess moderate consequences in all aspects. However, most of the likelihood fall under unlikely category. Hence, the risk posed is low. Proposed controls still need to be put in place to reduce the risk to lowest level.
The challenge to mimic the adhesive strength of naturally occuring mfps may be solved by producing rMAPs through fusion with peptide. First, mfps and tyrosinase are expressed in an E. coli expression system. Tyrosinase is used to convert tyrosine residue to dopa to imitate the the mfps found through natural extraction. Nevertheless, the tyrosinase-mediated post-translational modification onto rMAPs still have not met the quality of the adhesion (Castillo et al., 2017). Several adhesion tests will be performed to analyze the result of the fusion strategy.
There is a vast potential of improving the adhesiveness of mfps. Through this research, and further understanding of the combination of what works and not as well as mechanism of the mfps are obtained. Hopefully, this research will support future innovation in the field.
CASTILLO, J. J., SHANBHAG, B. K. & HE, L. 2017. Comparison of Natural Extraction and Recombinant Mussel Adhesive Proteins Approaches. In: PURI, M. (ed.) Food Bioactives: Extraction and Biotechnology Applications. Cham: Springer International Publishing.
CASTILLO, J. J. Q. 2017. Engineering Adhesive Proteins enhanced by self-assembling peptide and enzyme crosslinking. Master of Engineering, Monash University.
CHA, H. J., HWANG, D. S. & LIM, S. 2008. Development of bioadhesives from marine mussels. Biotechnology Journal, 3, 631-638.
CLARK, D. P. & PAZDERNIK, N. J. 2016. Chapter 10 – Recombinant Proteins. Biotechnology (Second Edition). Boston: Academic Cell.
FILPULA, D. R., LEE, S. M., LINK, R. P., STRAUSBERG, S. L. & STRAUSBERG, R. L. 1990. Structural and Functional Repetition in a Marine Mussel Adhesive Protein. Biotechnology Progress, 6, 171-177.
GANTAYET, A., OHANA, L. & SONE, E. D. 2013. Byssal proteins of the freshwater zebra mussel, Dreissena polymorpha. Biofouling, 29, 77-85.
GRIFFITHS, A. J., MILLER, J. H., SUZUKI, D. T., LEWONTIN, R. C. & GELBART, W. M. 2000. Making recombinant DNA. An Introduction to Genetic Analysis. 7th ed. New York: W. H. Freeman.
HALOULI, S., ASTHER, M., KRUUS, K., GUO, L., HAMDI, M., SIGOILLOT, J.-C. & LOMASCOIO, A. 2005. Characterization of a new tyrosinase from Pycnoporus species with high potential for food technological applications. Journal of Applied Microbiology, 98, 12.
HWANG, D. S., GIM, Y. & CHA, H. J. 2005. Expression of Functional Recombinant Mussel Adhesive Protein Type 3A in Escherichia coli. Biotechnology Progress, 21, 965-970.
HWANG, D. S., GIM, Y., YOO, H. J. & CHA, H. J. 2007a. Practical recombinant hybrid mussel bioadhesive fp-151. Biomaterials, 28, 3560-3568.
HWANG, D. S., SIM, S. B. & CHA, H. J. 2007b. Cell adhesion biomaterial based on mussel adhesive protein fused with RGD peptide. Biomaterials, 28, 4039-4046.
HWANG, D. S., YOO, H. J., JUN, J. H., MOON, W. K. & CHA, H. J. 2004. Expression of functional recombinant mussel adhesive protein Mgfp-5 in Escherichia coli. Applied and Environmental Microbiology, 70, 3352-3359.
HWANG, D. S., ZENG, H., MASIC, A., HARRINGTON, M. J., ISRAELACHVILI, J. N. & WAITE, J. H. 2010. Protein- and metal-dependent interactions of a prominent protein in mussel adhesive plaques. Journal of Biological Chemistry, 285, 25850-25858.
JEON, E. Y., HWANG, B. H., YANG, Y. J., KIM, B. J., CHOI, B.-H., JUNG, G. Y. & CHA, H. J. 2015. Rapidly light-activated surgical protein glue inspired by mussel adhesion and insect structural crosslinking. Biomaterials, 67, 11-19.
KITAMURA, M., KAWAKAMI, K., NAKAMURA, N., TSUMOTO, K., UCHIYAMA, H., UEDA, Y., KUMAGAI, I. & NAKAYA, T. 1999. Expression of a model peptide of a marine mussel adhesive protein in Escherichia coli and characterization of its structural and functional properties. Journal of Polymer Science Part A: Polymer Chemistry, 37, 729-736.
LIM, S., KIM, K. R., CHOI, Y. S., KIM, D.-K., HWANG, D. & CHA, H. J. 2011. In vivo post-translational modifications of recombinant mussel adhesive protein in insect cells. Biotechnology Progress, 27, 1390-1396.
NEWS, G. I. 2018. Postech Acad Industries Found Files Korean Patent Application for Ph-Responsive Nanoparticle Using Mussel Adhesive Protein for Drug Delivery and Method for Preparing the Same. Available: https://search-proquest-com.ezproxy.lib.monash.edu.au/docview/1986944479?accountid=12528&rfr_id=info%3Axri%2Fsid%3Aprimo.
NICKLISCH, S. C. T., DAS, S., MARTINEZ RODRIGUEZ, N. R., WAITE, J. H. & ISRAELACHVILI, J. N. 2013. Antioxidant efficacy and adhesion rescue by a recombinant mussel foot protein-6. Biotechnology Progress, 29, 1587-1593.
OHKAWA, K., NAGAI, T., NISHIDA, A. & YAMOMOTO, H. 2009. Purification of DOPA-Containing Foot Proteins from Green Mussel, Perna viridis, and Adhesive Properties of Synthetic Model Copolypeptides. The Journal of Adhesion, 85, 770-791.
PAPOV, V. V., DIAMOND, T. V., BIEMANN, K. & WAITE, J. H. 1995. Hydroxyarginine-containing polyphenolic proteins in the adhesive plaques of the marine mussel Mytilus edulis. Journal of Biological Chemistry, 270, 20183-20192.
RETTNER, R. 2017. DNA: Definition, Structure & Discovery [Online]. Live Science: Live Science. Available: https://www.livescience.com/37247-dna.html [Accessed 2018].
SCIENTIFIC, T. F. n.d.-a. Overview of Protein Expression [Online]. Thermo Fisher Scientific. Available: https://www.thermofisher.com/au/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-protein-expression-systems.html [Accessed 2018].
SCIENTIFIC, T. F. n.d.-b. Protein Expression Overview [Online]. Thermo Fisher Scientific. Available: https://www.thermofisher.com/au/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/protein-expression-handbook/pex-handbook-overview-protein-expression.html [Accessed 2018].
SCIENTIFIC, T. F. n.d.-c. Ribosomal Binding Site Sequence Requirements [Online]. Thermo Fisher Scientific. Available: https://www.thermofisher.com/au/en/home/references/ambion-tech-support/translation-systems/general-articles/ribosomal-binding-site-sequence-requirements.html [Accessed 2018].
SERVICE, U. F. N. 2017. WIPO PUBLISHES PATENT OF BENGT I. SAMUELSSON INSTITUTE OF LIFE SCIENCE RESEARCH FOR “MUSSEL ADHESIVE PROTEIN PRODUCT AND APPLICATION THEREOF FOR SUPPRESSING SOFT-TISSUE INFLAMMATION” (CHINESE INVENTOR). Available: https://search-proquest-com.ezproxy.lib.monash.edu.au/docview/1957124738?accountid=12528&rfr_id=info%3Axri%2Fsid%3Aprimo#.
SHUSTER, V. & FISHMAN, A. 2009. Isolation, Cloning and characterization of a Tyrosinase with Improved Activity in Organic Solvents from Bacillus megaterium. Journal of Molecular Microbiology and Biotechnology, 17, 13.
STEELE, E. n.d. Cell Fractionation: Definition, Steps & Methods [Online]. Study.com. Available: https://study.com/academy/lesson/cell-fractionation-definition-steps-methods.html [Accessed 2008].
TAMARIN, A., LEWIS, P. & ASKEY, J. 1976. The structure and formation of the byssus attachment plaque in Mytilus. Journal of Morphology, 149, 199-221.
WAITE, J. H. 1995. [1] Precursors of quinone tanning: Dopa-containing proteins. Methods in Enzymology. Academic Press.
WEEKLY, M. L. 2016. Patents; Patent Application Titled “Surface Immobilization of Various Functional Biomolecules Using Mussel Adhesive Protein” Published Online (USPTO 20160017279). Available: https://search-proquest-com.ezproxy.lib.monash.edu.au/docview/1762452829?accountid=12528&rfr_id=info%3Axri%2Fsid%3Aprimo.
WILKER, J. J. 2011. Redox and adhesion on the rocks. Nature Chemical Biology. Nature America Inc.
WILKER, J. J. 2015. Positive charges and underwater adhesion. sciencemag.org. AAAS.
WONG, H. H., KIM, Y. C., LEE, S. Y. & CHANG, H. N. 1998. Effect of post-induction nutrient feeding strategies on the production of bioadhesive protein in Escherichia coli. Biotechnology and Bioengineering, 60, 271-276.
XU, Y., ZHANG, H., JIANG, Q. & XIA, W. 2014. Preliminary purification and characterization of adhesive proteins from freshwater mussels. Journal of Adhesion, 90, 607-617.
YANG, B., KANG, D. G., SEO, J. H., CHOI, Y. S. & CHA, H. J. 2013. A comparative study on the bulk adhesive strength of the recombinant mussel adhesive protein fp-3. Biofouling, 29, 483-490.
ZENG, H., HWANG, D. S., ISRAELACHVILI, J. N. & WAITE, J. H. 2010. Strong reversible Fe<sup>3+</sup>-mediated bridging between dopa-containing protein films in water. Proceedings of the National Academy of Sciences, 107, 12850-12853.
ZHONG, C., GURRY, T., CHENG, A. A., DOWNEY, J., DENG, Z., STULTZ, C. M. & LU, T. K. 2014. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nature nanotechnology, 9, 9.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Marine Studies"
Marine studies look at the oceans and seas from the resources they provide, the diverse organisms and life that live in them to the threats and effects of climate change on marine habitats.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




