Improving Multiple Drug Resistance Cancer Treatment Efficacy
Info: 6525 words (26 pages) Dissertation
Published: 8th Jun 2021
Tagged: MedicinePharmacologyCancer
Microfluidic device for multi-drug resistance cancerous cell-derived exosomes isolation and the analysis of their response to nanotherapy
Abstract
Exosomes are membrane-enclosed phospholipid extracellular vesicles which were indicated as important biomarkers of cancerous cell functionality, such as multiple drug resistance (MDR). Nanoparticles based chemotherapy is a promising technology to overcome MDR by interfering the production and composition of exosomes. Therefore, tumor-derived exosomes post-treated by nanotherapy are implied to play critical roles of biomarkers on cancer MDR analysis. However, the efficient isolation of such exosomes from extracellular environment for their therapeutic response analysis remains challenging. In this study, we presented a microfluidic device consisting of exosome specific anti-CD63 immobilized degradable ciliated micropillars, which was capable to isolate the secreted exosomes from cell culture medium. The captured exosome can be recovered intact by degrading the ciliate of the micropillar using PBS soaking. Owing to the immobilization of antibody on the microfluidic device, nearly 70% of exosome from the biofluid could be isolated. So the secreted exosomes of the multi-drug resistance and ordinary human breast cancer cells pre-treated by free drug or nanotherapy could be isolated with high purity. Then the drug content of the isolated exosomes were measured to analysis of the MDR exosomal pathway response of MDR cells to different chemotherapeutic formations. Such analyses and further definition of the biomarkers of these exosomes could benefit the future investigations of accurately and reliably determine design principle, functional activity, and mechanisms of nanotherapy for multiple drug resistance overcoming.
Keywords Exosome · Microfluidic device· Isolation · nanoparticles therapy· Multi-drug resistance
1. Introduction
Nowadays, chemotherapy is playing an important role to against various cancers, such as breast cancer, ovarian cancer and lung cancer etc.(Fisher, Bryant, Wolmark et al. 1998, Le Chevalier, Arriagada, Le Pechoux et al. 2004, Parmar, Ledermann, Colombo et al. 2003) However, the efficiency of chemotherapy is mainly restricted by multiple drug resistance (MDR) acquiring by cancerous cells under long-time drug exposure (Foo and Michor 2014, Gottesman 2002). To overcome the MDR, new treatment methods are developed: one of the effective strategies is the introducing of nanotherapy which refer to technologies of delivering small chemotherapeutic agents by loading them into nanofomations such as nano-liposomes, polymeric micelles and other nanomateirals(Hu and Zhang 2012, Yuan, Cai, Xia et al. 2016, Zhang, Liu, Cui et al. 2017). The advantages of such strategy is obvious. Briefly, small molecule chemo drugs are transferred into cancerous cells via membrane translocators or passively diffuses, which make them easily to be refluxed back to the extracellular environment by the MDR cell membrane over-expressed P-glycoprotein (Pgp). Meanwhile, drug-loaded nanoparticles are internalized into cells through endocytosis pathways to overcome the MDR Pgp related effluxing mechanism (Bannunah, Vllasaliu, Lord et al. 2014, Oh and Park 2014). Thus, the involving of nanoparticles therapy (nanotherapy) into MDR cancer treatment did result in improved chemotherapy efficacy. However, for the further clinical translation and development of nanotherapy, it is essential to elaborate the specific process of nanoparticles on the MDR overcoming. The recent studies have shown that the enhanced cancer-derived exosome secreting pathway plays an important role for this improvement.(Soekmadji and Nelson 2015, Wang, Xu, Hua et al. 2016)
Exosomes are extra-cellular double-layer lipid vesicles with 40-120 nm in diameter secreted by the majority of viable cells to the bodily fluids, including blood, urine, and saliva. (Fevrier and Raposo 2004, van der Pol, Boing, Harrison et al. 2012, Yang, Weng, Mendrick et al. 2014). Typical exosomes sourced from epithelial cells, lymphoid cells, or tumor cells contents functional proteins, lipids and RNAs and act significant roles in intercellular communication (Kucharzewska and Belting 2013, Rak 2013) . Moreover, due to the universal existence and stability of exosomes in most of body fluid and resemblance of their contents to parental cells, exosomes have great potential to perform as traceable biomarkers for various diseases (Melo, Luecke, Kahlert et al. 2015, Tang and Wong 2015) . For example, the exosome derived by cancerous cells impact in many cancer-related pathways, such as tumor growth, metastasis, and angiogenesis (Kalluri 2016, Yang and Robbins 2011). Furthermore, cancer-derived exosomes have been found to participate in the MDR generating and acted as a major role in the process (Giallombardo, Taverna, Alessandro et al. 2016, Santos, Lima, Sarian et al. 2018). Corcoran et al. discovered exosomes contributed to docetaxel excretion of prostate cancer cells, and caused phenotypic change to non-resistant cell (Corcoran, Rani, O’Brien et al. 2012). Roohangiz et al. documented that the exosomes secreted by cisplatin-resistant cancer cells contained 2.5 times more cisplatin than of that from normal cancer cells (Safaei, Larson, Cheng et al. 2005) . These studies demonstrated the exosomal pathway effects through small drug efflux during the occurrences of MDR. However, the performance of exosomes in MDR overcoming nanotherapy and the interfering of nanoparticles on exosome generation and secretion still need to be evaluated (Oves, Qari, Felemban et al. 2018) .
To analyze the cancer-derived exosomes, it is important to develop the efficient technique to isolate the exosomes from body fluid. Different methods have been introduced. One of the conventional procedures includes the differential centrifugation which involves a series of centrifugation, filtration, and ultracentrifugation steps suffering from inconsistent exosome recovery rates, the breakage of exosome membrane and contamination of co-sedimentation of protein aggregates(Lamparski, Metha-Damani, Yao et al. 2002) Another procedure for exosome isolation is immuno-affinity capturing which applies the materials such as magnetic particles that are coated with a monoclonal antibody to specifically binds the exosome. This method has been implement as less time consuming and high specific exosome purity(Szatanek, Baran, Siedlar et al. 2015) Recently, research efforts have increased dramatically on developing immuno-affinity capturing microfluidic devices in order to isolate biological exosomes. Compare to the other procedure used as immuno-affinity capturing, microfluidic technology offers high isolation yield and functional integration for target exosomes(Kanwar, Dunlay, Simeone et al. 2014) Chen et al. reported an effective microfluidic exosome isolation platform, whose surface is immobilized with anti-CD63 antibody for capturing of exosomes from human serum(Chen, Skog, Hsu et al. 2010) Kanwar et al. developed a similar microfluidic device called ExoChip, which utilized antibody immobilized circular chamber to capture exosomes, following by fluorescent staining for on-chip isolation, quantification and characterization of circulating exosomes(Kanwar, Dunlay, Simeone et al. 2014) Therefore, antibody immobilized exosome isolation procedures were configured to enhance the capture efficiency. However, it is quite difficult to elute the captured exosomes from the antibody immobilized magnetic particles or the microfluidic device, which can present a major limitation of the possibilities for further experimentation on the extracted exosomes(Tauro, Greening, Mathias et al. 2012) .
Our group has presented a microfluidic device consisting of ciliated micropillars with fabricated porous 30–200 nm gap silicon nanowires through simply electroless etching using electro-deposited silver nanoparticles. This microfluidic device was able to preferentially physical trap exosome-like lipid vesicles by the porous silicon nanowire while allowing smaller proteins and larger nanoparticles with the size of cellular debris to pass through unhindered. Moreover, the captured liposome can be released by degrading the porous silicon nanowires in PBS buffer, thus allowing for the high integrity and purity of isolation contents recovery (Wang, Wu, Fine et al. 2013) However, this platform based on physical vesicle size isolation was restricted by surface lipid saturation limit. In this paper, the alternatively improvements and application of this microfluidic device are developed. The porous silicon nanowires of the device were fabricated by depositing gold nanopattern on the sidewalls of ciliated micropillars which is proved to be more effective and less time consuming. The nanowires of the ciliated micropillars were immobilized by the exosome-specific antibody, which allows efficient and non-invasive rapid isolation of specific exosomes with high purity. We hypothesized such improvements will significantly speed up the exosome isolation and analysis; benefit to acquire more accurate biological information. Moreover, The design of this microfluidic device provide the opportunities to isolate and quality the exosomes secreted from MDR cancerous cells and evaluate the interfering of drug-loaded nanoparticles on exosomes production and composition procedure. The outcome of this study would improve our understanding the mechanisms of exosomal responding to MDR profile during traditional chemotherapy or nanotherapy, thus leading to more efficient MDR cancer treatment.
2. Materials and Methods
2.1 Materials
Chloroauric acid (HAuCl4), (3-Aminopropyl)triethoxysilane(APTES), doxorubicin(Dox) and paclitaxel(PTX) were purchased from Sigma-Aldrich (St. Louis, MO). P-type (100) silicon wafers with resistivity of 0.005 Ω-cm were purchased from Silicon Quest (Santa Clara, CA). Polydimethylsiloxane (PDMS) and curing agent were obtained from Dow Corning (Midland, MI). Phosphate buffered saline (PBS, pH 7.2) was obtained from Gibco (Thermo-Fisher, Waltham, MA). Hydrofluoric acid was from Honeywell International Inc. N-maleimidobutyryl-oxysuccinimide ester (GMBS) and thiol functionalized streptavidin were purchased from Nanocs Inc (Woburn, MA). The biotinylated anti-CD63 antibody and control IgG was purchased from Abcam Co.(Boston, MA). Human breast cancer cell line MDA-MB-231 cells were purchased from the American Type Culture Collection (Rockville, MD, USA). The multidrug resistant MDA-MB-231/MDR cell line was cultured in our lab. All the cells were cultured in DMEM (Thermo-Fisher, Waltham, MA) supplemented with 10 % FBS. The exosome Labelling Kit (Exo-GlowTM) was purchased from System Bioscience (Palo Alto, CA). Amphiphilic copolymer methoxy-poly(ethylene glycol)-poly(ε-caprolactone) (mPEG-PCL, Mw: 5000:5000) was purchased from Polymer Source Inc.(Canada)
2.2 Device fabrication
The pillars on the microfluidic device were prepared as previously described (Wang, Wu, Fine et al. 2013). Silicon wafer with micropillars was first dipped into HF solution to remove native oxide. Then the wafer is assembled with the Teflon electroplating cell, and sealed with a Viton O-ring and aluminum foil. A platinum mesh immersed into 0.5 M HAuCl4 plating solution was applied as electrode. The reverse pulse electroplating is performed to deposit uniform gold nanopattern on the sidewalls of the micropillars. After that, the wafer was chemically etched in a mixture of 0.1M H2O2 and 2.9M HF solution for 2 minute to get nanowire on the ciliated micropillar. The Au nanopattern was then stripped by immersing into gold etchant (Transene Inc., Denvers, MA) for 2 minutes. The substrate was then rinsed in excess water, ethanol and dried by blow air.
2.2 Device construction
The micropillars on the wafer were first functionalized with the APTES by immersing into 0.1M H2O2 at 70°C overnight subsequently in 1% APTES isopropanol solution at 80°C for 24h as reported (Gunda, Singh, Norman et al. 2014). And the polydimethylsiloxane (PDMS) gaskets were made in a 10:1 elastomer base-curing agent ratio and incubated at 70°C for 4 hours under vacuum followed by cleaning with isopropanol deionized water. The APTES functionalized wafer and PDMS gaskets were treated with oxygen plasma at 0.2 torr for 30 seconds. Then the wafer and PDMS gasket were manually bonded and subsequently baked at 37°C for 10 min to get the microfluidic device based on the cillated micropillars.
2.3 Surface modification
For antibody immobilizing, the device was rinsed in the 0.1mg/ml GMBS solution for 30 min followed by washing with purified water. Then the microfluidic device was sequential dipped into thiol functionalized streptavidin and biotinylated anti-CD63 antibody PBS solution at 4°C. The antibody immobilized microfluidic device was obtained after washing by 1% BSA solution and PBS. The control IgG immobilized microfluidic device was achieved as the similar method.
2.4 Exosome isolation and recovery
The cells were cultured DMEM supplemented with 10 % exosome depleted FBS (System Bioscience Palo Alto, CA) for 12 h. Then the culture medium were collected and centrifuged at 2000 rpm for 10 min to remove cell debris. The exosome in the medium was labelled by Exo-Green Exosome Labelling Kit according to protocol. The labelling exosome solution was continuously injected into the anti-CD63 or control IGG immobilized microfluidic device through the inlet at a flow rate of 10μL/min by the Fusion 200 high precision syringe pump (Chemyx, Inc.,Stafford, TX). The waste was then collected at the outlet. The fluoresce intensity of inlet and outlet exosome was measured by BD plate reader (BD Biosciences, Billerica, MA). The isolation efficiency of exosome was calculated by the ratio fluoresce intensity value of inlet and outlet. For exosome imaging, PBS solution was injected into the device to wash untapped impurity. The exosomes in the device were fixed by 0.5 × Karnovsky’s fixative and then sequencing dipped with water, 25% ethanol, 50% ethanol, 70% ethanol and 100% ethanol for 10 min each. The exosome on the micropillars of microfluidic device were examined using FEI Nova Nano scanning electron microscope with 2 nm gold-palladium layer and Nikon A1 Confocal Laser Microscopy Imaging System with 488 nm excitation (Melville, NY, USA).
To recovery the isolated exosome from micropillar, the microfluidic device channels were immersing by PBS for 8 h and then flushed by PBS. The obtained the exosomes in PBS was placed on a mica slide and observed by Multimode Atomic Force Microscope with tapping mode (Bruker, Billerica, MA).
2.5 The preparation of drug loading nanoparticles
The doxorubicin encapsulated liposome (Doxoves®) was purchased from FormuMax Scientific, Inc. (Sunnyvale, CA). The paclitaxel containing micellar nanoparticles was prepared by co-solvent evaporation method. Briefly, mPEG-PCL (10 mg) and 1 mg of PTX were dissolved in 0.2 mL acetone. This solution was added to 1 mL of water. The mixture was then sonicated in ice bath for 2 min. The excess organic solvent was removed under vacuum. The micellar nanoparticles solution was filtered through a 0.22 μm Minisart syringe filter (Sartorius, Germany) to remove free PTX and subsequently lyophilized. The PTX content in the nanoparticles was measured by Agilent 1200 High Performance Liquid Chromatography System (HPLC) (Agilent, Santa Clara, CA).
2.6 Muti-drug resistance characterization
MDA-MB-231/MDR and MDA-MB-231 were seeded in a 96 well plate at 5×103 cells/well for 24 h. Then the cell culture medium was replaced with new medium containing free Dox, Doxoves®, free PTX (dissolved in DMSO) or PTX-micellar nanoparticles with different concentration for culturing another 72 h. The chemo-sensitivity of the MDA-MB-231/MDR and MDA-MB-231 cells were measured by MTT assay as literature reported (Napierska, Thomassen, Rabolli et al. 2009)
2.7 Cell treatment and exosome analysis
MDA-MB-231/MDR and MDA-MB-231 were pretreated with free Dox, Doxoves®, free PTX or PTX-micellar nanoparticles by a fixed drug concentration of 40 µg/mL for 4h. Then the medium were discarded and the cells washed twice with cold PBS and incubated with DMEM supplemented with 10 % exosome depleted FBS for another 12 h. The culture mediums were collected and centrifuged at 2000 rpm for 10 min to remove the cell debris. For exosome isolation, the collected mediums were injected into the anti-CD63 immobilized microfluidic device through the inlet at a flow rate of 10μL/min and washed by PBS twice. Then the isolation exosomes were collected by immersed the micropillars into PBS containing 0.1% Triton X-100 for 8h and the channels were flushed by PBS containing 0.1% Triton X-100 which increase the drug solubility. The drug content in the obtained exosomes was tested by Agilent 1200 High Performance Liquid Chromatography System.
Results and discussion
Design of the anti-CD63 antibody immobilized microfluidic device
A microfluidic device which is capable to filter the target fluid with simultaneously multidimensional is highly desirable to fulfill the mission to isolate the exosomes. We assigned the similar multidimensional structure microfluidic device structure as we reported previously.(Fig. S1) (Wang, Wu, Fine et al. 2013) As illustrated in Fig. 1A, the microfluidic device we prepared consists of a PDMS caps and a concentric ellipses-flow fluidic channel containing antibody immobilized ciliated hexagonally packs of micropillars with 2.6 µm in diameter and 15 µm in heights. The gaps among the pillars are designed as 900 nm for the fluid flowing. A circular inlet and outlet were also connected to the micropillar area with a syringe pump for sample introduction and extraction. We hypothesized that the interstitial sites of the micropillars capable of physically (1) bypassing the unneeded small sample components, such as small molecular protein and micron-size cellular debris; (2) filtering the larger sample components, such as cells. Moreover, the antibody immobilized nanowire forest on the sidewalls of ciliated micropillars are dedicated to selectively capture the exosomes flowing through the device interstitial sites. (Fig. 1B) And the porous nanowires could be dissolved in buffers to integrity isolate the captured exosomes for their further analysis. Thus, such microfluidic device is capable to apply for the MDR cancerous cell-derived exosomes isolation and the analysis of their response to nanotherapy.
In order to enhance exosome capturing efficiency, the sidewalls nanowires forests is essential to increase the contacting area between micropillars and bypassing fluid. The nanowire was fabricated by the combination of electroplating, electroless metal-assisted etching techniques. First, by the governing of the electrochemical reaction and reactant diffusion, as well as the properties of the reactants and silicon substrates, a uniform gold nanopattern is deposited onto the sidewalls of micropillars (Fig. 2A). Second, the uniform nanowire arrays were obtained by subsequently etching the micropillar surface and stripping the excess gold nanopartterns(Fig. 2B). Finally, the sillicon surface of the device was functionalized of amine groups followed by decorating of streptavidin, thus the biotinylation anti-CD63 antibody could be stable immobilized on the nanowires of the ciliated micropillars for exosome capturing (Fig. 2C).
Isolation of exosome by microfluidic device
Fluorescein-labelled cancer cell-derived exosomes were labelled and used as the traceable samples to verify the functionality and isolation efficacy of our antibody immobilized microfluidic device. The samples was injected to the inlet through the PDMS cap and extracted at the outlet. After that, the captured exosome by the ciliated micropillars on the microfluidic device was observed by SEM. As shown in Fig. 3A&B, the captured exosomes are visualized on the sidewalls of micropillars and trapped in the porous of the nanowire, which demonstrated the capacity of microfluidic device with ciliated micropillars to capture the exosomes from body fluid. To quantity of the efficacy on exosomes isolation, the eluting outlet samples were collected and the fluorescence intensity was divided versus that of the totals injected samples. The unspecific IgG immobilized microfluidic device was used as control. As Fig.3C shown, 75% of the total exosome retention was observed by the anti-CD63 immobilized microfluidic device, while only less than 20% of exosome retention rate was shown for no-selective IgG immobilized one. The functionalization of antibodies against CD63, an antigen commonly overexpressed in exosomes greatly contributes to this significant efficacy of exosome capturing (Szatanek, Baran, Siedlar et al. 2015)(28) The captured exosome by the antibody immobilized microfluidic device was further visualized by confocal laser microscopy. As shown in Fig. 4A and B, the continuously green fluoresce dots which represent the captured exosomes clusters were observed to absorb on the sidewalls of micropillars. Notably, the three-dimension imaging showed the majority of the exosome was trapped around the micropillars instead of the bottom of the microfluid device, which demonstrated the forming of turbulence among the micropillars enhanced the absorbing chances of exosome and anti-CD63 leading the high isolation efficacy. These revealed results showed that our microfluidic device with anti-CD63 immobilized ciliated micropillars offers obvious advantages on exosome isolating.
The recovering of captured exosomes
It has been reported that the degradation of porous silicon nanowires in the presence of PBS buffer(Chiappini, Liu, Fakhoury et al. 2010) Thus, the microfluidic device prototype we developed is not only applied for exosomes capturing but also recovering the captured exosomes through the degradation of the porous nanowire of the ciliated micropillar. The degradation of nanowire was accomplished by soaking the channels of microfluidic device with PBS for 8 h after capturing exosomes. As shown in Fig. 5A, the nanowires of ciliated micropillars in the microfluidic device were fully disappeared, which lead the stripping of the anti-CD63 immobilizing layer and trapping sites to release the capturing exosomes. Commonly, the traditional methods of isolating exosomes always interfere with the structure of the exosomes and hardly to show the morphology of native exosomes (Szatanek, Baran, Siedlar et al. 2015) However, the recovered exosome from our microfluidic device maintained its 100 nm sphere morphology and the integrity of surface structure as characterized by Atom Force Microscopy (Fig.5B). Overall, the microfluidic device we designed presents the significantly capacity on high purity exosomes selectively isolating and recovering from biofluid.
Measuring of drug content of isolated exosomes
Intrinsic or acquired multiple drug resistance (MDR) which considerably limits the efficacy of chemotherapeutics, is mediated by different mechanisms, such as P-gP effluxing (Hayeshi, Masimirembwa, Mukanganyama et al. 2006, Kim, Haney, Zhao et al. 2016) Recently, nanotechnology holds great promising development for drug delivery systems with controlled-release and drug-resistance overcoming profiles for MDR cancer treatment(Hayeshi, Masimirembwa, Mukanganyama et al. 2006) It is reported that the altering biophysical properties of anticancer drugs by nanoparticles conjugation or encapsulation was able to bypass the recognition of P-gP(Brigger, Dubernet and Couvreur 2012) Moreover, the paramount overlap of exosome expelling and the uptake of nanoparticles are still unclear. Especially considering that the secreting of cancerous cell exosomes plays critical roles in drug resistance generating and transferring (Soekmadji and Nelson 2015, Wang, Xu, Hua et al. 2016), the effects of exosomal pathway related to the drug-loaded nanoparticles MDR cellular internalization, further to the MDR overcoming need to be well evaluated.
As the results above, our microfluidic device with antibody immobilized cilliary micropilars is a well-suited platform to be used for effective and non-destructive exosomes isolation. By isolating exosomes from MDR cells treated with nanotherapy through this prototype, the drug of nanoformation efflux profile in the exosomal pathway can be determined, which lead a better understanding of drug expulsion mechanism through the exosomal pathway and circumventing MDR effect. For this purpose, two different of drug-loaded nanoformations were prepared: the commercialized doxorubicin-loaded liposome (Doxoves) and synthetized paclitaxel-loaded mPEG-PCL micellar nanoparticle. The normal human breast cancer cell MDA-MB-231 and its multiple drug resistance type MDA-MB-231/MDR were both treated by free small drugs and nanotherapeutic formations. After that, the expelled exosomes in the cultured mediums were isolated through anti-CD63 immobilized microfluidic device and the total drug contents in the exosomes was measured to study exosomal pathway corresponding to different forms of drug treatment. As shown in Figure 5A & B, both the exosomes from nanoparticles treated non-resistant and resistant cell showed much lower drug content than free drug treated counterpart. Moreover, it is notably that free drug treated MDR cells showed more drug efflux than its counterpart sensitive cell due to the MDR profiles ( approximately 2 times and 1.5 times higher of DOX and PTX, respectively), while nanoformations treated MDR cells showed comparable drug efflux than its sensitive counterpart. Considering the same treated drug concentration on both sensitive/MDR cancer cells, these results demonstrated the existing overlap of the cellular exosomal expelling pathway and bypassing P-gP effluxing effect of nanoparticles. However, the enhanced cellular internalization of different of nanoformations were not inducing the actual MDR overcoming (Fig. S2). The slow drug releasing from the nanoparticles or other mechanisms related MDR such as anti-apoptosis may be responsible to the nanotherapy insensitivity of cancerous cells. (Hu and Zhang 2009, Notarbartolo, Cervello, Dusonchet et al. 2002) Thus, developing a novel drug-loaded nanoplatform not only internalizing the cancerous cells bypassing exosomal effluxing pathways but also owning intracellular burst release profiles and personalizing capacity against the multi-mechanisms of multiple drug resistance is essential for the future clinical applications of nanotherapy.
Conclusion
We developed a microfluidic device with surface nanowire modified ciliated micropillars. By the immobilization of anti-CD63 on the nanowire, this device was capable of the specifically capturing the exosomes in the biofluid, while simultaneously bypassing smaller proteins/cell debris and filtering larger particles such as cells. The captured exosomes were able to be recovered without structure destructive by dissolving the porous nanowires in buffers. Therefore, this microfluidic device could be used as a rapidly and efficiently tool to selectively isolate exosomes from complex biological samples while maintaining their structural integrity, offering potential benefit to a range of both diagnostic and therapeutic applications. This device was further applied for the isolating of exosomes derived by drug-loaded nanoparticles treated multiple drug resistance cancerous cells. Due to the superior performance of the microfluidic device, the relationships between the exosomal pathway and the MDR-related drug expulsion under the nanotherapy were identified. These results insure the future studies of MDR biomarkers on secreted exosomes under varies treatment procedure, as which will contribute to our further mechanism understandings on MDR acquiring and overcoming.
Reference
Bannunah, A. M., D. Vllasaliu, J. Lord, et al. (2014). “Mechanisms of Nanoparticle Internalization and Transport Across an Intestinal Epithelial Cell Model: Effect of Size and Surface Charge.” Mol Pharmaceut 11(12): 4363-4373.
Brigger, I., C. DubernetP. Couvreur (2012). “Nanoparticles in cancer therapy and diagnosis.” Adv Drug Deliver Rev 64: 24-36.
Chen, C., J. Skog, C. H. Hsu, et al. (2010). “Microfluidic isolation and transcriptome analysis of serum microvesicles.” Lab Chip 10(4): 505-511.
Chiappini, C., X. W. Liu, J. R. Fakhoury, et al. (2010). “Biodegradable Porous Silicon Barcode Nanowires with Defined Geometry.” Adv Funct Mater 20(14): 2231-2239.
Corcoran, C., S. Rani, K. O’Brien, et al. (2012). “Docetaxel-Resistance in Prostate Cancer: Evaluating Associated Phenotypic Changes and Potential for Resistance Transfer via Exosomes.” Plos One 7(12).
Fevrier, B.G. Raposo (2004). “Exosomes: endosomal-derived vesicles shipping extracellular messages.” Curr Opin Cell Biol 16(4): 415-421.
Fisher, B., J. Bryant, N. Wolmark, et al. (1998). “Effect of preoperative chemotherapy on the outcome of women with operable breast cancer.” J Clin Oncol 16(8): 2672-2685.
Foo, J.F. Michor (2014). “Evolution of acquired resistance to anti-cancer therapy.” J Theor Biol 355: 10-20.
Giallombardo, M., S. Taverna, R. Alessandro, et al. (2016). “Exosome-mediated drug resistance in cancer: the near future is here.” Ther Adv Med Oncol 8(5): 320-322.
Gottesman, M. M. (2002). “Mechanisms of cancer drug resistance.” Annu Rev Med 53: 615-627.
Gunda, N. S. K., M. Singh, L. Norman, et al. (2014). “Optimization and characterization of biomolecule immobilization on silicon substrates using (3-aminopropyl)triethoxysilane (APTES) and glutaraldehyde linker.” Appl Surf Sci 305: 522-530.
Hayeshi, R., C. Masimirembwa, S. Mukanganyama, et al. (2006). “The potential inhibitory effect of antiparasitic drugs and natural products on P-glycoprotein mediated efflux.” Eur J Pharm Sci 29(1): 70-81.
Hu, C. M. J.L. F. Zhang (2012). “Nanoparticle-based combination therapy toward overcoming drug resistance in cancer.” Biochemical Pharmacology 83(8): 1104-1111.
Hu, C. M. J.L. F. Zhang (2009). “Therapeutic Nanoparticles to Combat Cancer Drug Resistance.” Current Drug Metabolism 10(8): 836-841.
Kalluri, R. (2016). “The biology and function of exosomes in cancer.” J Clin Invest 126(4): 1208-1215.
Kanwar, S. S., C. J. Dunlay, D. M. Simeone, et al. (2014). “Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes.” Lab Chip 14(11): 1891-1900.
Kim, M. S., M. J. Haney, Y. Zhao, et al. (2016). “Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells.” Nanomedicine 12(3): 655-664.
Kucharzewska, P.M. Belting (2013). “Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress.” J Extracell Vesicles 2.
Lamparski, H. G., A. Metha-Damani, J. Y. Yao, et al. (2002). “Production and characterization of clinical grade exosomes derived from dendritic cells.” J Immunol Methods 270(2): 211-226.
Le Chevalier, T., R. Arriagada, C. Le Pechoux, et al. (2004). “Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer.” New Engl J Med 350(4): 351-360.
Melo, S. A., L. B. Luecke, C. Kahlert, et al. (2015). “Glypican-1 identifies cancer exosomes and detects early pancreatic cancer.” Nature 523(7559): 177-182.
Napierska, D., L. C. J. Thomassen, V. Rabolli, et al. (2009). “Size-Dependent Cytotoxicity of Monodisperse Silica Nanoparticles in Human Endothelial Cells.” Small 5(7): 846-853.
Notarbartolo, M., M. Cervello, L. Dusonchet, et al. (2002). “Resistance to diverse apoptotic triggers in multidrug resistant HL60 cells and its possible relationship to the expression of P-glycoprotein, Fas and of the novel anti-apoptosis factors IAP (inhibitory of apoptosis proteins).” Cancer Letters 180(1): 91-101.
Oh, N.J. H. Park (2014). “Endocytosis and exocytosis of nanoparticles in mammalian cells.” Int J Nanomed 9: 51-63.
Oves, M., H. A. Qari, N. M. Felemban, et al. (2018). “Exosomes: A Paradigm in Drug Development against Cancer and Infectious Diseases.” J Nanomater.
Parmar, M. K. B., J. A. Ledermann, N. Colombo, et al. (2003). “Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial.” Lancet 361(9375): 2099-2106.
Rak, J. (2013). “Extracellular vesicles – biomarkers and effectors of the cellular interactome in cancer.” Frontiers in Pharmacology 4.
Safaei, R., B. J. Larson, T. C. Cheng, et al. (2005). “Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells.” Mol Cancer Ther 4(10): 1595-1604.
Santos, J. C., N. D. Lima, L. O. Sarian, et al. (2018). “Exosome-mediated breast cancer chemoresistance via miR-155 transfer.” Sci Rep-Uk 8.
Soekmadji, C.C. C. Nelson (2015). “The Emerging Role of Extracellular Vesicle-Mediated Drug Resistance in Cancers: Implications in Advanced Prostate Cancer.” Biomed Res Int.
Szatanek, R., J. Baran, M. Siedlar, et al. (2015). “Isolation of extracellular vesicles: Determining the correct approach (Review).” Int J Mol Med 36(1): 11-17.
Tang, M. K.A. S. Wong (2015). “Exosomes: Emerging biomarkers and targets for ovarian cancer.” Cancer Lett 367(1): 26-33.
Tauro, B. J., D. W. Greening, R. A. Mathias, et al. (2012). “Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes.” Methods 56(2): 293-304.
van der Pol, E., A. N. Boing, P. Harrison, et al. (2012). “Classification, Functions, and Clinical Relevance of Extracellular Vesicles.” Pharmacol Rev 64(3): 676-705.
Wang, X. H., C. F. Xu, Y. T. Hua, et al. (2016). “Exosomes play an important role in the process of psoralen reverse multidrug resistance of breast cancer.” J Exp Clin Canc Res 35.
Wang, Z. X., H. J. Wu, D. Fine, et al. (2013). “Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles.” Lab Chip 13(15): 2879-2882.
Yang, C.P. D. Robbins (2011). “The roles of tumor-derived exosomes in cancer pathogenesis.” Clin Dev Immunol 2011: 842849.
Yang, X., Z. Q. Weng, D. L. Mendrick, et al. (2014). “Circulating extracellular vesicles as a potential source of new biomarkers of drug-induced liver injury.” Toxicol Lett 225(3): 401-406.
Yuan, Y. L., T. G. Cai, X. Xia, et al. (2016). “Nanoparticle delivery of anticancer drugs overcomes multidrug resistance in breast cancer.” Drug Deliv 23(9): 3350-3357.
Zhang, M., E. G. Liu, Y. N. Cui, et al. (2017). “Nanotechnology-based combination therapy for overcoming multidrug-resistant cancer.” Cancer Biol Med 14(3): 212-227.
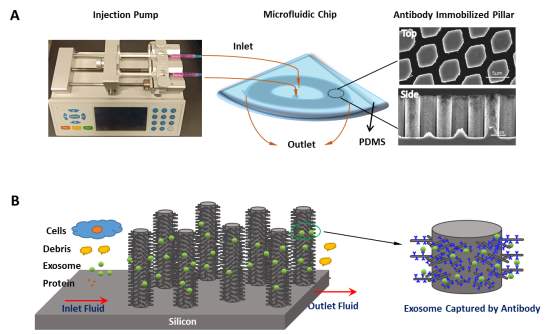
Fig. 1 Schematic of the antibody immobilized micropillars array for exosome isolation. (A) Experimental set-up of the microfluidic device system for exosomes isolation.
(B) The exosomes are captured by antibody immobilized micropillars when bypassing the microfluidic system.
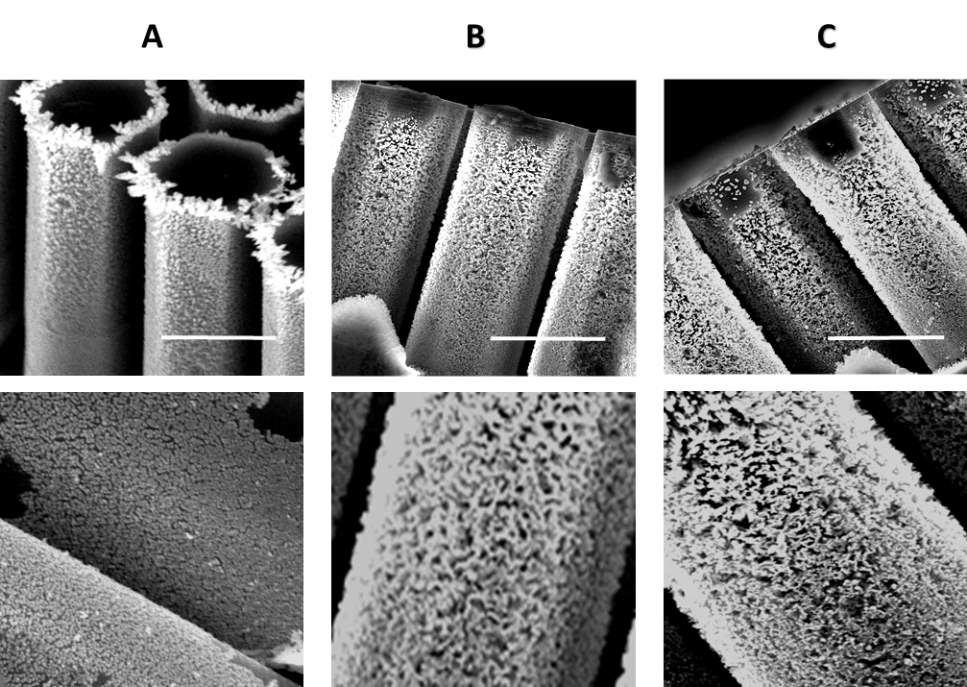
Fig. 2 SEM images of the antibody immobilized micropillars preparation stages. (A)Uniform gold deposited on silicon micropilliars (top) and its close-up view (bottom). (B) Porous silicon nanowires are formed on the sidewalls of the micropillars after etching (top) and its close-up view (bottom). (C) Anti-CD63 antibody were immobilized on the porous micropillars(top) and its close-up view (bottom). All scale bars are 5 µm.
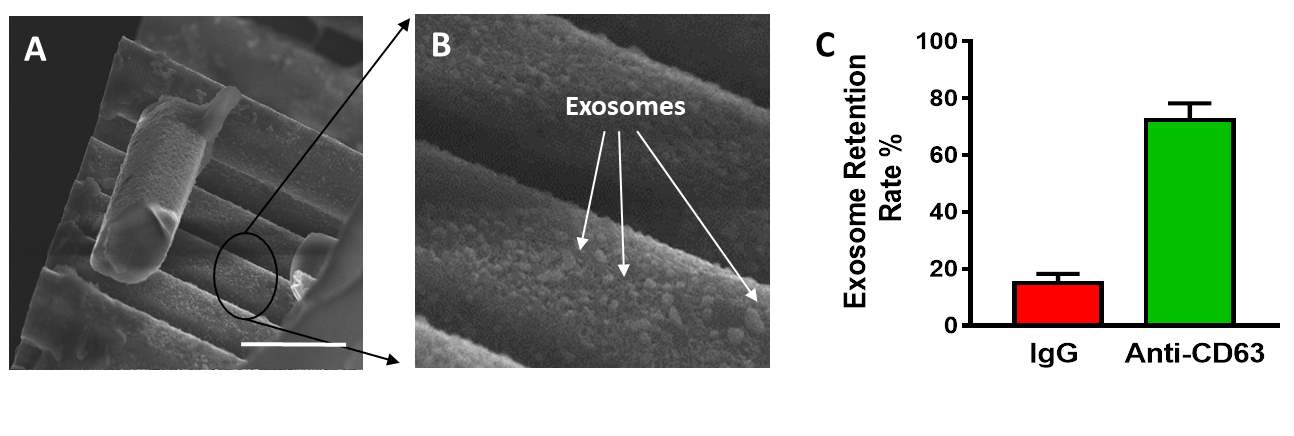
Fig. 3 Exosome retention by the antibody immobilized micropillar array. (A) SEM images of exosome captured by micropillars(Scale bar 5 µm). (B) Close-up view of captured exosome on the sidewall of micropillar (Scale bar 500 nm). (C) Comparison of exosome retention rate of anti-CD63 antibody immobilized micropillar and IgG immobilized micropillar
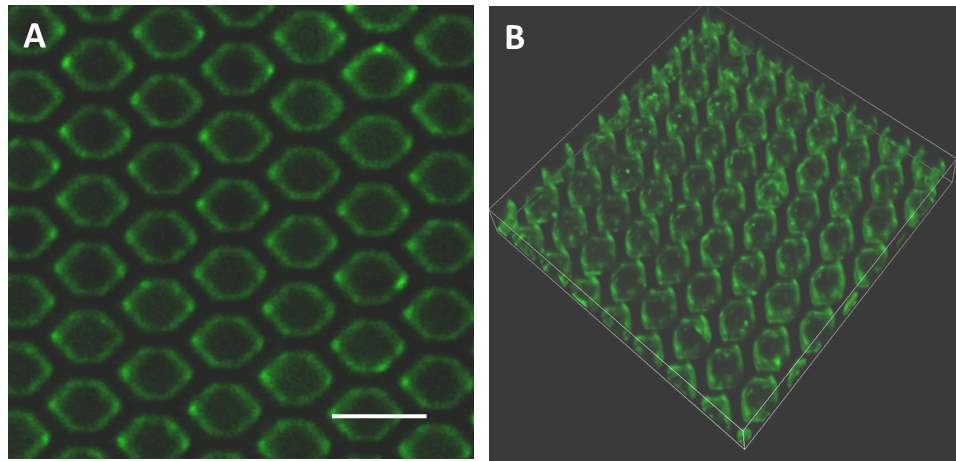
Fig.4 Confocal microscopy images of fluorescein-labelled exosomes captured by (A) micropillar and (B) its 3D imaging. Scale bars represent 10 µm.
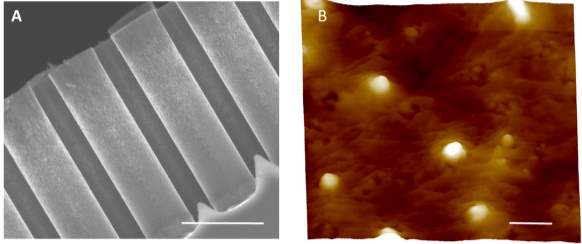
Fig. 5 (A) SEM image of the micropillars after soaking in PBS buffer to remove the captured exosome. (Scale bar represents 5 µm) (B) Atom forced microscopy image of the recovery exosome from micropillars. (Scale bar represents 200 nm)
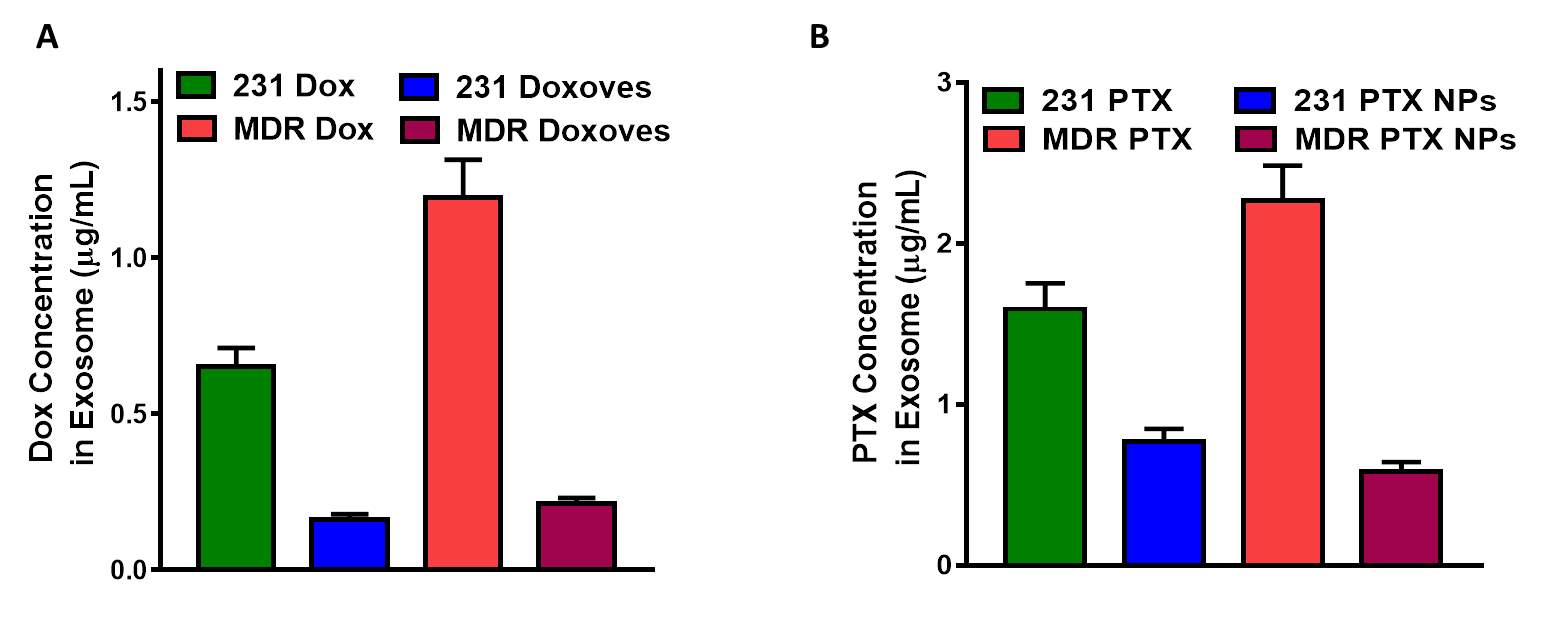
Fig. 6 (A) Doxorubicin and Paclitaxel content in the exosomes isolated from MDA-MB-231 human breast cancer cells (231) and multi-drug resistance MDA-MB-231 human breast cancer cells (MDR) after treating by different drug formations.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Cancer"
Cancer is a disease in which cells grow or reproduce abnormally or uncontrollably. Cancerous cells have the potential to spread to other areas of the body in a process called metastasis.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




