Genetic Polymorphisms and Platelet Reactivity in the Early Phases of Acute Coronary Syndromes
Info: 9939 words (40 pages) Dissertation
Published: 18th Feb 2022
Tagged: Medical
GENETIC POLYMORPHISMS OF ABCB1, CYP2C19, CYP3A5, CYP4F2 AND PLATELET REACTIVITY IN THE EARLY PHASES OF ACUTE CORONARY SYNDROMES
Abstract
Aims
The aim was to study 7 polymorphic markers of genes encoding proteins involved in the absorbtion, metabolism and pharmacokinetics of clopidogrel among patients with an acute coronary syndrome (ACS).
Materials and Methods
81 ACS and PCI patients older than 18 years treated with dual antiplatelet therapy. Platelet function testing and ABCB1, CYP2C19, CYP3A5, CYP4F2 genotyping were performed. Predictive role of categorical variables such as genotypes (carriers and non-carriers of polymorphism) on platelet reactivity (PRU, PI) was assessed by logistic regression (for categorical outcomes) and linear regression (for continuous outcomes) analysis. A p-value
Results
Regarding clopidogrel response, 62 patients (76,5 %) were clopidogrel responders and 19 were non-responders (23,5 %). Mean PRU value and the percentage of platelet inhibition were 170,0 ± 50,9 PRU and 28,6 ± 19,9 %, respectively. The effects of CYP2C19*2 polymorphisms on PRU (166,0 ± 50,8 vs. 190,7 ± 48,2, p CYP2C19*1/*2 + CYP2C19*2/*2 (16,2% vs. 53,8% p 0,05). By the logistic regression analysis, CYP2C19*2 (OR: 4,365, CI: 1,25 – 17,67, p = 0,022) was independent predictor of HPR at CYPC19*17 – 34,9 ± 25,0 vs. 25,9 ± 16,9 (OR=0,32; р=0,006), the rest of polymorphisms had no influence.
Conclusions
We found that on-clopidogrel platelets reactivity in the early phase of ACS is influenced primarily by CYP2C19 polymorphisms. We believe that the findings of the present study could supply additional evidence regarding the clinical appropriateness of the CYP2C19 genetic testing for tailoring antiplatelet therapy in the early phase of ACS.
Keywords: clopidogrel, ticagrelor, prasugrel, acute coronary syndrome, pharmacogenetics, cytochrome P450, polymorphism, percutaneous coronary intervention
Introduction
As a standard of care for patients with acute coronary syndrome undergoing percutaneous coronary intervention, dual antiplatelet therapy is routinely used [1]. The P2Y12 inhibitor clopidogrel is a prodrug, therefore it needs to be metabolized in the liver, which makes its pharmacokinetics dependent on the activity of the enzymes involved. For clopidogrel, these enzymes being CYP2C19, CYP2B6, CYP1A2 for transformation of the prodrug into 2-oxo-clopidogrel as stage one, and CYP2C19, CYP2B6, CYP3A4, CYP3A5, CYP2C9 to transform that intermediate product into an active metabolite as stage two [2]. Not only does metabolism of clopidogrel involve a two-stage transformation, but also tiny transporter P-glycoproteins to facilitate prodrug absorbtion at the apical membranes of the intestinal cells. This P-glycoprotein is encoded by the ABCB1 gene. As a result, there are numerous sites for a possible breakdown which lead to altered response to clopidogrel. These causes for altered clopidogrel response could be divided into three groups: pharmacokinetic markers like intestinal absorbtion, metabolic activation in the liver and pharmacodynamic markers. These causes have a clear link to variants of the genes coding the enzymes involved, so called polymorphisms. These polymorphisms which are either suspected or proven causes are mentioned in this article as follows: ABCB1 C3435T, ABCB1 C>T, rs4148738 for P-glycoprotein; CYP2C19*17 C-806T, CYP2C19*2 681G>A, CYP2C19*3 636G>A, CYP3A5*3 A6986G for genes CYP2C19 and CYP3A5 of the cytochrome P450 enzymes family; CYP4F2 C(Val433Met)T as a pharmacodynamic marker for CYP4F2 enzyme which is also responsible for epoxyeicosatrienoic acids metabolism [3].
This altered response to clopidogrel poses a challenge to physician and could potentionally lead towards such complications as stent thrombosis or bleeding [4,5]. Although, studies suggest that the presence of polymorphisms does affect laboratory clopidogrel resistance, the clinical implications for routine genetic testing are not stated according to the current guidelines [6]. This is so because the “critical mass” of the studies showing that altered response to clopidogrel might seriously jeopardize the safety of percutaneous coronary intervention and cause stent thrombosis or bleeding has not been achieved yet.
One of the way to assess the response to clopidogrel is to measure platelet reactivity with the VerifyNow P2Y12 Assay. The therapeutic range is over 85 but less than 208 platelet reactivity units (PRUs) [7]. While PRU level under 85 may be a predictor of a bleeding event, PRU level over 208 is a sign that the higher clopidogrel reactivity is present and a physician might consider taking action. This is the time for a personalized approach. There were some trials which assessed doubling and tripling the dose of clopidogrel for low responders [8,9]. However, the current strategy for low responders is switching to another P2Y12 inhibitor like ticagrelor or prasugrel [10].
Studies have shown that polymorphisms are more frequent in Asians than in Caucasians [11] Therefore, a physician might consider measuring platelet reactivity for such a patient or switch to another P2Y12 inhibitor.
Materials and methods
Study population
The study protocol was approved by the Ethics Committee of Russian Medical Academy of Continuous Professional Education, Ministry of Health of the Russian Federation, Moscow, and all patients gave written informed consent for participation. It was conducted in accordance with the Declaration of Helsinki and consistent with applicable guidelines for good clinical practice. Patients included in this study were recruited from October 2014 to September 2015 in Moscow city clinical hospital №1, Moscow, Russian Federation. Those enrolled 81 ACS and PCI patients older than 18 years were treated with dual antiplatelet therapy, 100 mg of aspirin daily and either a 300 or 600 mg loading dose following 75 mg maintenance dose daily of clopidogrel. Postprocedural therapy consisted of 100 mg aspirin once daily and 75 mg clopidogrel once daily. The major exclusion criteria included high risk of bleeding, thrombocytopenia, contraindications to aspirin or clopidogrel, malignancies, pregnancy, and cerebrovascular events within the past 3 months. These factors included age, sex, smoking, diabetes mellitus, hypertension (BP ≥ 140/90 mmHg), hyperlipidemia, bodymass index, family history of coronary artery disease, previous myocardial infarction, and coronary stents were obtained from patient’s files or interview.
Blood sampling
A total of two blood samples from all 81 patients were collected on the 3-5th day after the last dose of clopidogrel taken for genotyping and platelet reactivity. Blood (2 ml each) for DNA analysis was sampled from the peripheral vein using ethylene diamine tetra acetate (EDTA) tubes (VACUETTE® (Greiner Bio-One, Austria) and stored at −80 °C. Blood samples (2 ml each) were taken in tubes with 3.2% sodium citrate for the measurement of platelet activity in response to ADP at 4 h after the last dose of clopidogrel taken [12].
Extraction of peripheral blood DNA
Genomic DNA was extracted from peripheral blood by using “DNA ‑ EKSTRAN-1” (ZAO Syntol, Russia) according to the manufacturer’s protocol. The quantitative concentration of DNA was measured by the Nanodrop Spectrophotometer (ND-2000, USA).
ABCB1, CYP2C19, CYP3A5 and CYP4F2 genotyping
A panel of 7 SNP of ABCB1 (C3435T, rs1045642), ABCB1 (C>T, rs4148738), CYP2C19*2 (681G>A, rs4244285), CYP2C19*3 (636G>A, rs4986893), CYP2C19*17 (C-806T, rs1224856), CYP3A5*3 (A6986G, rs776746), CYP4F2 (C>T, Val433Met, rs2108622) was selected based on previous investigations [12-14]. Base numbering and allele definitions follow the nomenclature of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee (www.cypalleles.ki.se). The genotypes were determined with a TaqMan® Single Nucleotide Polymorphism Genotyping Assay kit and a TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions, using an ABI PRISM® Sequence Detector 7000 (Applied Biosystems). All the PCR reactions were carried out in a 10-μL reaction volume containing genomic DNA 15 ng, oligonucleotide primers 0.5 pM, 1 μL 10 PCR buffer, deoxynucleotides (dNTPs) 250 μM, magnesium chloride 3 mM, and DNA polymerase 0.25 U. The cycling program involved preliminary denaturation at 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 60 s, and elongation at 72°C for 60 s, followed by a final elongation step at 72°C for 7 min [15].
Platelet function assay
Platelet function tests were performed within 1 h of sample acquisition utilizing VerifyNow P2Y12 assay (Accumetrics, San Diego, CA, USA) within 5 days after clopidogrel loading (early phase). The glycoprotein IIb/IIIa inhibitors and intravenous anticoagulants other than unfractionated heparin were not approved for use in patients with ACS in this study. The level of platelet aggregation is expressed in P2Y12 reactivity unit (PRU) and in platelet percentage inhibition (PI). The assessment was performed within 1 hour after venous blood collection. The VerifyNow system is a rapid point-of-care platelet function test system. The VerifyNow P2Y12 test measures ADP-induced platelet aggregation and reports results as P2Y12 Reaction Units (PRU). A second channel in the test device activates platelets through the thrombin receptor pathway, which is P2Y12-receptor-independent and provides a simultaneous estimate of baseline total platelet function, which is reported as “Base PRU”. The percent inhibition of ADP-induced platelet aggregation is calculated from the PRU and Base PRU values. A cutoff value for higher platelet reactivity (HPR) was defined as PRU > 208 in the present study, despite the lack of consensus on the cutoff value associated with clinical outcomes [16]. The reference values were PRU values 208 (high OTR to clopidogrel or clopidogrel non-responders). Moreover, a cutoff for lower platelet reactivity (LPR) was defined as PRU
Statistical analysis
Qualitative variables were expressed as relative and absolute frequencies and quantitative variables as the mean (M) ± standard deviation (SD). The Kolmogorov Smirnov test was used to assess normality. Continuous variable comparisons were performed using the Student T test and analysis of variance. The chi-square test and analysis of variance (ANOVA) were used to compare variables between the subgroups, and the Fisher exact test was used when any expected cell count was
Results
Characteristics of the study population and genotyping
A total of 81 Caucasian patients were included in the study. In 81 ASC patients, 77 received PCI, and 4 received standard medical treatment without PCI. There were no significant differences in the demographic, clinical, and laboratory findings between the study subgroups according to platelet reactivity levels (Table 1).
Table 1. Baseline demographics and clinical characteristics of the study cohort with resistant to clopidogrel (PRU > 208) and a normal response to clopidogrel (PRU
| Characteristic | Total
(n=81) |
Patients with
PRU |
Patients with
PRU > 208 (n=19) |
р |
| Age, years mean ± SD | 63,9±10,9 | 63,2±11,0 | 66,2±10,4 | 0,265 |
| Men, n (%) | 64 (79,0) | 50 (80,6) | 14 (73,7) | 0,360 |
| BMI, mean ± SD | 27,8±3,1 | 28,0±3,2 | 27,8±3,1 | 0,361 |
| Sub-type ACS, n (%) | ||||
| UA | 10 (12,3) | 9 (14,5) | 1 (5,3) | 0,290 |
| STEMI | 50 (61,7) | 35 (56,5) | 15 (78,9) | |
| NSTEMI | 17 (21,0) | 14 (22,6) | 3 (15,8) | |
| Indeterminate | 4 (4,9) | 4 (6,5) | 0 (0,0) | |
| Risk factors, n (%) | ||||
| DM | 16 (19,8) | 12 (19,4) | 4 (21,1) | 0,552 |
| HBP | 75 (92,5) | 58 (95,1) | 17 (89,5) | 0,340 |
| Current smoking status | 17 (21,5) | 20 (0,0) | 5 (26,3) | 0,385 |
| Dyslipidemia | ||||
| Prior MI | 14 (17,2) | 8 (12,9) | 6 (31,5) | 0,497 |
| Prior stroke | 5 (6,2) | 4 (6,5) | 1 (5,6) | 0,686 |
| Prior PCI | 12 (14,8) | 8 (12,9) | 4 (21,1) | 0,295 |
| CHF II-III FC | 3 (3,7) | 1 (1,6) | 2 (10,5) | 0,136 |
| Arrhythmia | 8 (9,9) | 4 (6,5) | 4 (21,1) | 0,083 |
| Stent trombosis | 2 (2,5) | 1 (1,7) | 1 (5,3) | 0,416 |
| Drug-eluting stent | 30 (38,9) | 25 (43,1) | 5 (26,3) | 0,331 |
| Involved vessel | ||||
| LM | 4 (4,9) | 4 (6,5) | 0 (0,0) | 0,335 |
| LAD | 39 (49,4) | 32 (53,3) | 7 (36,8) | 0,161 |
| LCX | 27 (34,2) | 22 (36,7) | 5 (26,3) | 0,295 |
| LMA | 4 (4,9) | 3 (4,8) | 1 (5,3) | 0,665 |
| RCA | 23 (28,3) | 15 (24,6) | 8 (42,1) | 0,115 |
| Stent diameter, mm | 2,75±6,8 | 2,6±0,8 | 3,0±0,3 | 0,049 |
| Stent length, mm | 25,5±5,1 | 26,3 (10,9) | 22,7 (6,4) | 0,178 |
| Medication, n (%) | ||||
| Aspirin | 81 (100) | 62 (100) | 19 (100) | – |
| Statin | 81 (100) | 62 (100) | 19 (100) | – |
| CCB | 5 (6,2) | 2 (3,2) | 3 (15,8) | 0,081 |
| Diuretics | 22 (27,2) | 17 (27,4) | 5 (26,3) | 0,588 |
| PPI | 80 (98,8) | 62 (100) | 18 (94,7) | 0,235 |
| Beta-blockers | 71 (89,9) | 54 (90,0) | 17 (89,5) | 0,621 |
| ACE-inhibitors | 62 (76,5) | 49 (79,0) | 13 (68,4) | 0,254 |
ACS, acute coronary syndrome; STEMI, ST-segment elevation myocardial infarction; NSTEMI, non-ST-segment elevation myocardial infarction; UA, unstable angina; CHF, chronic heart failure; FC – functional class; ACE – angiotensin converting enzyme inhibitor; BMI – body mass index; CCB, calcium channel blocker; DM, diabetes mellitus; HBP, hypertension; IM, intermediate metabolizer; LAD, left anterior descending artery; LCX, left circumflex artery; LM, left main coronary artery; LMA, left marginal artery; RCA, right coronary artery; MI, myocardial infarction; PCI, percutaneous coronary intervention; PM, poor metabolizer; PPI, proton pump inhibitor. Student’s t-test for continuous variables and χ2 test for categorical variables, p
All patients were successfully genotyped for 7 genetic variants, ABCB1 (C3435T, rs1045642), ABCB1 (C>T, rs4148738), CYP2C19*2 (681G>A, rs4244285), CYP2C19*3 (636G>A, rs4986893), CYP2C19*17 (C-806T, rs1224856), CYP3A5*3 (A6986G, rs776746), CYP4F2 (C>T, Val433Met, rs2108622)(Table 2). Apart from ABCB1 (C3435T, rs1045642) and CYP3A5*3 (A6986G, rs776746), no significant deviations from Hardy‑Weinberg equilibrium were observed for all of the genetic variants (Table 2).
Table 2. Genotype frequencies and Hardy-Weinberg equilibrium test.
| Allele | Genotype, n (%) | MinorAllele (%) | HWE* | ||||
| Homozygous noncarriers | Heterozygous carriers | Homozygous carriers | χ2 | p | |||
| ABCB1 (rs1045642) | Obs. | CC 17 (21,0%) | CT 51 (63,0%) | TT 13 (16,0%) | 47,5 | 5,57 | 0,018 |
| Exp. | 22,3 | 40,4 | 18,3 | ||||
| ABCB1 (rs4148738) | Obs. | CC 15 (18,5%) | CT 45 (55,6%) | TT 21 (25,9%) | 53,7 | 1,11 | 0,291 |
| Exp. | 17,4 | 40,3 | 23,4 | ||||
| CYP2C19*2 (rs4244285) | Obs. | GG 68 (84,0%) | AG 13 (16,0%) | AA 0 (0,0%) | 8,0 | 0,62 | 0,432 |
| Exp. | 68,5 | 12,0% | 0,5 | ||||
| CYP2C19*3 (rs4986893) | Obs. | GG 81 (100%) | AG 0 (0,0%) | AA 0 (0,0%) | 0,0 | – | – |
| Exp. | – | – | – | ||||
| CYP2C19*17 (rs1224856) | Obs. | CC 57 (70,4%) | CT 23 (28,4%) | TT 1 (1,2%) | 25,4 | 0,62 | 0,428 |
| Exp. | 57,9% | 21,1 | 1,9 | ||||
| CYP3A5*3 (rs776746) | Obs. | GG 78 (96,3%) | AG 3 (3,7%) | AA 0 (0,0%) | 1,35 | 11,9 | 0,001 |
| Exp. | 77,1 | 4,8 | 0,1 | ||||
| CYP4F2*3 (rs2108622) | Obs. | CC 40 (49,4%) | CT 38 (46,9%) | AA 3 (3,7%) | 27,1 | 2,79 | 0,095 |
| Exp. | 43,0 | 32,0 | 6,0 | ||||
*HWE – Hardy-Weinberg equilibrium test Obs.- observed genotype Exp.- expected genotype
Effects of ABCB1, CYP2C19, CYP3A5 and CYP4F2 genotypes on the P2Y12-antiplatelet effect of clopidogrel
Regarding clopidogrel response, 62 patients (76,5 %) were clopidogrel responders and 19 were non-responders (23,5 %). Mean PRU value and the percentage of platelet inhibition were 170,0 ± 50,9 PRU and 28,6 ± 19,9 %, respectively. There were no significant differences in Base PRU levels among any genotypes. At ABCB1, CYP2C19, CYP3A5 and CYP4F2 genotypes on P2Y12-reactivity unit (PRU) values and the percentage of platelet inhibition (PI) after ACS with PCI (ANOVA, U-test) are shown in Table 3.
The effects of CYP2C19*2 polymorphisms on PRU (166,0 ± 50,8 vs. 190,7 ± 48,2, p CYP2C19*1/*2 + CYP2C19*2/*2 (16,2% vs. 53,8% p 0,05).
Table 3. Influence of ABCB1, CYP2C19, CYP3A5 and CYP4F2 genotypes on P2Y12-reactivity unit (PRU) values and the percentage of platelet inhibition (PI) after ACS with PCI (ANOVA, U-test)*.
| Genotype | PRU value, mean±SD | p | Percentage inhibition (PI) % | p |
| ABCB1 | ||||
| rs1045642 | ||||
| CC | 187,0 ± 26,6 | 0,123 | 20,0 ± 13,1 | 0,077 |
| CT + TT | 165,5 ± 54,9 | 30,9 ± 20,8 | ||
| rs4148738 | ||||
| CC | 169,2 ± 52,8 | 0,946 | 29,2 ± 17,7 | 0,657 |
| CT + TT | 170,1 ± 50,9 | 28,6 ± 19,9 | ||
| CYP2C19 | ||||
| rs4244285 | ||||
| GG | 166,0 ± 50,8 | 0,038 | 30,6 ± 20,0 | 0,013 |
| GA + AA | 190,7 ± 48,2 | 18,1 ± 16,3 | ||
| rs4986893 | ||||
| GG | 170,0 ± 50,9 | – | 28,6 ± 19,9 | – |
| GA + AA | – | – | ||
| rs1224856 | ||||
| CC | 176,8 ± 44,1 | 0,064 | 25,9 ± 16,9 | 0,076 |
| CT + TT | 153,8 ± 62,3 | 34,9 ± 25,0 | ||
| CYP3A5 | ||||
| rs776746 | ||||
| GG | 169,6 ± 51,8 | 0,723 | 28,7 ± 20,3 | 0,696 |
| GA + AA | 180,3 ± 18,0 | 26,0 ± 4,0 | ||
| CYP4F2 | ||||
| rs2108622 | ||||
| CC | 174,8 ± 51,0 | 0,407 | 27,7 ± 18,3 | 0,902 |
| CT + TT | 165,3 ± 51,0 | 29,5 ± 21,5 |
*Analysis of variance (ANOVA) was used to calculate difference in PRU value and Mann – Whitney U-test was used to calculate difference in Percentage inhibition
By the logistic regression analysis, CYP2C19*2 (OR: 4,365, CI: 1,25 – 17,67, p = 0,022) was independent predictor of HPR at
The linear regression was used to calculate the odds ratio of changes in on-clopidogrel platelet reactivity (PRU and PI) between carriers and non-carriers of the minor allelic variant: for PRU – CYPC19*2 – 190,7 ± 48,2 vs. 166,0 ± 50,8 (OR=0,27; р=0.026) and CYPC19*17 – 153,8 ± 62,3 vs. 176,8 ± 44,1 (OR=0,3; р=0,01); for PI – 18,1 ± 16,3 vs. 30,6 ± 20,8 (OR=0,33; р=0,005) and CYPC19*17 – 34,9 ± 25,0 vs. 25,9 ± 16,9 (OR=0,32; р=0,006), the rest of polymorphisms had no influence.
Linkage disequilibrium analysis and Haplotype analysis
We used “SNPStats” for the analysis of linkage disequilibrium and haplotype analysis (http://bioinfo.iconcologia.net/en/SNPStats_web) [17]. The pairs of polymorphisms are considered to be in “strong LD” if the one-sided upper 95% confidence bound on D′ is ≥0.98 and the lower bound is ≥0.70. The ABCB1 rs1045642 and ABCB1 rs4148738 polymorphisms showed a strong linkage disequilibrium (D’ = 0,72 and D’ = 0,99, relatively). On the other hand, the polymorphisms in the CYP2C19 gene (*2 (rs4244285) and *17 (rs1224856)) were in low linkage disequilibrium (D’ = 0,36) (p
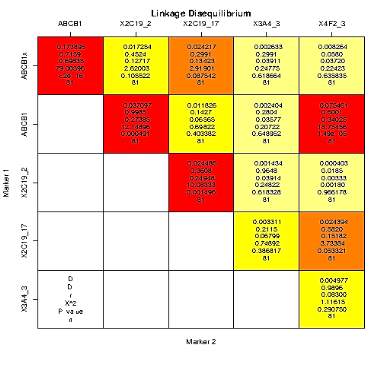
Figure 1. Linkage disequilibrium analysis of ABCB1, CYP2C19, CYP3A5 and CYP4F2 polymorphisms.
Table 4. Haplotype association with P2Y12-reactivity unit (PRU) and percentage of platelet inhibition (PI) after ACS with PCI for all SNP. (n=81, crude analysis)
| ABCB1 | CYP | Haplotype analysis results | ||||||
| (rs1045642) | (rs4148738) | CYP 2C19*2 (rs4244285) | 2C19*17 (rs1224856) | 3A5*3 (rs776746) | 4F2*3 (rs2108622) | Haplotype freq.* | PRU mean difference (95% CI) | P |
| PRU | ||||||||
| T | C | G | C | G | C | 0.3011 | 0.00 | — |
| T | C | G | C | G | T | 0.0336 | -60.65 (-92.21 – 29.08) | 0.00023 |
| PI % | ||||||||
| T | C | G | C | G | C | 0.3146 | 0.00 | — |
| C | T | A | T | G | C | 0.0313 | -20.91 (-37.14 – 4.67) | 0.013 |
| T | T | G | T | G | T | 0.0122 | 46.29 (23.1 – 69.48) | 0.00014 |
| T | C | G | C | G | T | 0.0062 | 68.2 (35.59 – 100.81) | |
* Freq. ‑ frequency
Table 5. Haplotype association with P2Y12-reactivity unit (PRU) and percentage of platelet inhibition (PI) after ACS with PCI for all SNP, except CYP2C19*17. (n=81, crude analysis).
| ABCB1 | CYP | Statistics | |||||
| (rs1045642) | (rs4148738) | 2C19*2 (rs4244285) | 3A5*3 (rs776746) | 4F2*3 (rs2108622) | Freq. | Difference (95% CI) | P |
| PRU | |||||||
| T | C | G | G | C | 0.3373 | 0.00 | — |
| C | T | G | G | C | 0.2124 | -13.8 (-26.74 – -0.86) | 0.038 |
| T | C | G | G | T | 0.0498 | -47.35 (-78.03 -16.67) | 0.0029 |
| PI % | |||||||
| T | C | G | G | T | 0.0259 | 31.74 (9.79 – 53.7) | 0.0051 |
* Freq. ‑ frequency
Table 6. Haplotype association with percentage of platelet inhibition (PI) after ACS with PCI for all CYP polymorphisms. (n=81, crude analysis; the P2Y12-reactivity unit (PRU) had no association).
| CYP | Statistics | |||||
| 2C19*2 (rs4244285) | 2C19*17 (rs1224856) | 3A5*3 (rs776746) | 4F2*3 (rs2108622) | Freq. | Difference (95% CI) | P |
| G | C | G | C | 0.5514 | 0.00 | — |
| G | T | G | C | 0.1007 | 12.88 (1.44 – 24.31) | 0.03 |
| G | T | G | T | 0.0112 | 50.02 (19.15 – 80.89) | 0.002 |
* Freq. ‑ frequency
Discussion
In general, 81 ACS patients undergoing PCI were included in this study at T, rs4148738), CYP2C19*2 (681G>A, rs4244285), CYP2C19*3 (636G>A, rs4986893), CYP2C19*17 (C-806T, rs1224856), CYP3A5*3 (A6986G, rs776746) can impact clopidogrel pharmacokinetics and pharmacodynamics [19] in both healthy volunteers and ACS patients. The polymorphisms of CYP4F2 (C>T, Val433Met, rs2108622) genes selected in this study have no impact on clopidogrel pharmacokinetics, but this gene coding cytochrome P450 4F2 subfamily, enzyme which takes part in biosynthesis of epoxyeicosatrienoic acids – important vasoactive products of arachidonic acid metabolism with a wide range of biological actions in the cardiovascular system, including influence on platelet reactivity.
Simultaneously, there was a paucity of data regarding the combined effect of several ABCB1, CYP2C19, CYP3A5 and CYP4F2 at-risk alleles, and insufficient data to measure the linkage disequilibrium analysis and haplotype analysis. We found that CYP2C19*2 increases the on-clopidogrel platelet reactivity which is associated with a higher risk of ischemic cardiovascular events [20]. Furthermore, we observed that CYP2C19*17 was associated with lower platelet reactivity than non-carriers. In the previous studies with the Russian patients enrolled (AL Komarov. [21], E.Z. Golukhova. [22], Mazurov A.V. [23], Mirzaev KB [24], Galyavich AS [25], Sumarokov A.V. [26]) was investigated the role of polymorphism CYP2C19, CYP2C19, ABCB1 genes and other risk factors of the poor clopidogrel antiplatelet effect.
For example, Komarov A.L. et al. showed that male sex, low ejection fraction, multivascular lesion of coronary arteries, the ABCB1 C3435T polymorphism and proton pump inhibitors intake significantly increase the risk of clopidogrel non-responsiveness, and CYP2C19*2 carriers had a 2.4-times increased risk of thrombotic cardiovascular complications (95% CI=1.2‑4.9; р=0.01) [21]. Frequency of both alleles CYP2C19*2 and CYP2C19*17 among current ACS patients was close to that observed among Russian patients from Moscow region, from Northern, Central, and Eastern Siberia in other previously published studies with ACS patients.
None of the other polymorphisms was associated with differences in the results of the 5-day platelet reactivity analysis by VerifyNow P2Y12 assay.
According to linkage disequilibrium analysis, the ABCB1 rs1045642 and ABCB1 rs4148738 polymorphisms showed a strong linkage disequilibrium (D’ = 0,72 and D’ = 0,99, relatively). In contrast, the polymorphisms in the CYP2C19 gene (*2 (rs4244285) and *17 (rs1224856)) were in low linkage disequilibrium (D’ = 0,36) (p
There are about 34 CYP2C19 alleles including CYP2C19*2 and CYP2C19*3, CYP2C19*17. The most common CYP2C19 polymorphisms are CYP2C19*2, CYP2C19*3, and CYP2C19*17 [27]. Carriers of CYP2C19*2 and CYP2C19*3 polymorphisms are poor metabolizers (PM), i.e. they have decreased activity of liver enzymes and reduced clopidogrel biotransformation. Carriers of CYP2C19*17 polymorphism are ultra-rapid metabolizers (UM). About 50% of the Mongoloid, 34% of Negroid, and 18% of Caucasians are CYP2C19*2 carriers [3,28]. CYP2C19*3 allele frequency is less than 1% in Caucasians and Negroid and less than 7% in Mongoloids [29]. CYP2C19*17 was found in 25.7% of Germans [30], 22% of Norwegians [31] and less than 4% of Asian population (Korean, Japanese, Chinese) [32]. CYP2C19*2 was shown to be a strong determinant of ischemic cardiovascular events in Asian patients, whereas the relation between CYP2C19*2 allele and the magnitude of risk has been inconsistent in Caucasian [3, 29, 33, 34].
Consequently, the evaluation of the interindividual differences in the prevalence of CYP2C19 gene polymorphisms is very important in the countries such as the Russian Federation because of the high multiethnicity. According to the results obtained in the previous studies, from 16% up to 27.5% of patients with ACS in different regions of Russia have at least one CYP2C19*2 allele variant affecting clopidogrel metabolism. For example, the results of the previous studies in both healthy volunteers and ACS patients demonstrated that 38.0% of Kalmyks, 23.0% of Russians, 20.0% of Tatars, 18.0% of Chechens, 12.0% of Ingushes, 18.8% of Carachay, 14.0% of Circassian, 12.7% of Avar, 14.5% of Lak, 5.0% of Dargin, 23% of Yakuts, 21% of Buryats, 15% of Altayans, and of 15% Tuvinians populations carried at least one CYP2C19*2 allele variant [35-41].
According to the association between CYP2C19 and clopidogrel pharmacokinetics, numerous studies have confirmed an important role of CYP2C19 loss-of-function alleles in HPR, determined by ADP-induced platelet reactivity in CAD patients [3,9,42-48]. In addition, some studies have also revealed that CYP2C19*17 (increased activity allele) results in enhanced platelet inhibition and possibly an increased bleeding risk [3,49,50]. Several clinical studies have detected a significant association between CYP2C19*2 allele and adverse cardiovascular events including an increased risk for stent thrombosis [3,12,14,29,42,44,51,52]. Furthermore, studies revealed that the usage of CYP2C19-genotyping among patients treated with clopidogrel leads to a larger overall benefit compared to lower risk indications, such as medical management of ACS and stable CAD [53,54]. Therefore, the influence of CYP2C19 on clinical outcomes has been most evident among ACS with PCI patients. Reported meta-analyses suggest that clopidogrel-treated ACS/PCI patients who are CYP2C19*2 carriers have an increased risk compared to CYP2C19*1/*1 patients for main adverse cardiovascular events [55].
In fact, it is very difficult to perform large randomized studies guided by genotype-directed treatment. In previous studies CYP2C19 genotype-directed antiplatelet therapy trials have been reported (CLOVIS-2 , ELEVATE TIMI-56 , RAPID-GENE ,RAPID STEMI, ACCEL-AMI-2C19 , ACCEL-2C19, GeCCO, PAPI-2, GIANT, TAILOR-PCI, NCT01097343). For example, In RAPID GENE trials it was determined that CYP2C19 genetic testing after ACS with PCI can be done effectively for genotype-directed treatment [56], which subsequently was expanded in the RAPID STEMI trial [57]. Although the reported “pharmacodynamiс” randomized studies support CYP2C19 genotype-directed antiplatelet therapy, clinical trials powered for actual clinical outcomes are ultimately more likely to definitively establish or refute the clinical utility of CYP2C19 genotyping.
The American College of Cardiology Foundation (ACCF), the American Heart Association (AHA), the Society for Cardiovascular Angiography and Interventions, the Society of Thoracic Surgeons and European Society of Cardiology (ESC) did not recommend routine genetic testing prior to clopidogrel administration, because they lack sufficient evidence. However, they did suggest consideration of genetic testing for patients of high risk of adverse cardiovascular events (high-risk PCI procedures, previous stent thrombosis, DM etc.) and that CYP2C19 poor metabolizers should be prescribed an alternative antiplatelet regimen [58]. Similarly, the Royal Dutch Association for the Advancement of Pharmacy Pharmacogenetics Working Group (KNMPPWG), and the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommend genetic testing and genotype-directed treatment for high-risk patients [59]. The CPIC guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy: (1) standard dosing of clopidogrel is warranted among patients with a predicted CYP2C19 EM or UM phenotype (i.e., *1/*1, *1/*17, and *17/*17); if the patient is a carrier of CYP2C19 PM (i.e., *2/*2, *2/*3, *3/*3), current literature supports the use of an alternative antiplatelet agent (e.g., prasugrel, ticagrelor) [60].
There are some limitations to this study. Firstly, it includes a relatively small number of patients. Secondly, the current study does not evaluate the association of the clinical outcome and the genetic polymorphism.
Conclusions
Seven polymorphic markers of genes encoding proteins involved in the absorption (ABCB1), metabolism, pharmacokinetics (CYP2C19, CYP3A5), and (CYP4F2) of clopidogrel in 81 ACS patients were studied. We found that on-clopidogrel platelet reactivity in the early phase of ACS is influenced primarily by CYP2C19 polymorphisms. Another ADP-inhibitors, such as ticagrelor and prasugrel may increase the degree of platelet inhibition [3,60], but several big randomized trials have failed to confirm clinical efficacy of tailoring antiplatelet therapy according to the results of platelet function tests [61,62]. We believe that the findings of the present study could supply additional evidence regarding the clinical appropriateness of the CYP2C19 genetic testing for tailoring antiplatelet therapy in the early phase of ACS. Nevertheless, to draw definite conclusions on this problem, further large-scale researches are essential.
Disclosure
This study has been supported by Russian Science Foundation ‑ project 16‑15-00227 “Fundamental research and exploratory research conducted in the top priority areas”.
References
1. Valgimigli M, Bueno H, Byrne RA et al. 2017 ESC focused update on dual antiplatelet therapy in coronary artery disease developed in collaboration with EACTS. European journal of cardio-thoracic surgery : official journal of the European Association for Cardio-thoracic Surgery 53(1), 34-78 (2018).
2. Simon T, Verstuyft C, Mary-Krause M et al. Genetic determinants of response to clopidogrel and cardiovascular events. The New England journal of medicine 360(4), 363-375 (2009).
3. Mega JL, Close SL, Wiviott SD et al. Cytochrome p-450 polymorphisms and response to clopidogrel. The New England journal of medicine 360(4), 354-362 (2009).
4. Konova OD, Mirzaev KB, Ryzhikova KA, Sozaeva ZA, Andreev DA, Sychev DA. The Association of the rs2108622 Single Nucleotide Polymorphism in the CYP4F2 Gene with Resistance to Clopidogrel. Clinical Therapeutics 39(8), e62
5. Mirzaev KB, Rytkin E, Grishina EA et al. CRT-100.09 The Impact of CYP2C19, ABCB1Genes Polymorphisms and CYP3A4 Isoenzyme Activity on the Incidence of Stent Implantation Complications for Patients With an Acute Coronary Syndrome. JACC: Cardiovascular Interventions 10(3 Supplement), S6 (2017).
6. Levine GN, Bates ER, Bittl JA et al. 2016 ACC/AHA Guideline Focused Update on Duration of Dual Antiplatelet Therapy in Patients With Coronary Artery Disease. A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines 68(10), 1082-1115 (2016).
7. Tantry US, Bonello L, Aradi D et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. Journal of the American College of Cardiology 62(24), 2261-2273 (2013).
8. Mega JL, Hochholzer W, Frelinger AL et al. Dosing clopidogrel based on CYP2C19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. Jama 306(20), 2221-2228 (2011).
9. Collet JP, Hulot JS, Anzaha G et al. High doses of clopidogrel to overcome genetic resistance: the randomized crossover CLOVIS-2 (Clopidogrel and Response Variability Investigation Study 2). JACC. Cardiovascular interventions 4(4), 392-402 (2011).
10. Aradi D, Storey RF, Komocsi A et al. Expert position paper on the role of platelet function testing in patients undergoing percutaneous coronary intervention. European heart journal 35(4), 209-215 (2014).
11. Sychev DA, Shuev GN, Suleymanov SS et al. Comparison of CYP2C9, CYP2C19, CYP2D6, ABCB1, and SLCO1B1 gene-polymorphism frequency in Russian and Nanai populations. Pharmacogenomics and personalized medicine 10 93-99 (2017).
12. Harmsze A, Van Werkum JW, Bouman HJ et al. Besides CYP2C19*2, the variant allele CYP2C9*3 is associated with higher on-clopidogrel platelet reactivity in patients on dual antiplatelet therapy undergoing elective coronary stent implantation. Pharmacogenetics and genomics 20(1), 18-25 (2010).
13. Shalia KK, Shah VK, Pawar P, Divekar SS, Payannavar S. Polymorphisms of MDR1, CYP2C19 and P2Y12 genes in Indian population: effects on clopidogrel response. Indian heart journal 65(2), 158-167 (2013).
14. Simon T, Verstuyft C, Mary-Krause M et al. Genetic determinants of response to clopidogrel and cardiovascular events. The New England journal of medicine 360(4), 363-375 (2009).
15. Kaneko A, Lum JK, Yaviong L et al. High and variable frequencies of CYP2C19 mutations: medical consequences of poor drug metabolism in Vanuatu and other Pacific islands. Pharmacogenetics 9(5), 581-590 (1999).
16. Malinin A, Pokov A, Spergling M et al. Monitoring platelet inhibition after clopidogrel with the VerifyNow-P2Y12(R) rapid analyzer: the VERIfy Thrombosis risk ASsessment (VERITAS) study. Thrombosis research 119(3), 277-284 (2007).
17. Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics 22(15), 1928-1929 (2006).
18. Nagashima Z, Tsukahara K, Morita S et al. Platelet reactivity in the early and late phases of acute coronary syndromes according to cytochrome P450 2C19 phenotypes. Journal of cardiology 62(3), 158-164 (2013).
19. Jiang XL, Samant S, Lesko LJ, Schmidt S. Clinical pharmacokinetics and pharmacodynamics of clopidogrel. Clinical pharmacokinetics 54(2), 147-166 (2015).
20. Buonamici P, Marcucci R, Migliorini A et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. Journal of the American College of Cardiology 49(24), 2312-2317 (2007).
21. Komarov AL, Shakhmatova ОО, Ilyushenko TA et al. Assessment of cardiovascular complications risk in patients with stable ACS treated with klopidogrel: platelet function or genetic testing? Doktor.Ru. 6: 11-19. (2012)
22. Golukhova EZ, Grigorjan MV, Rjabinina MN et al. The platelet reactivity after percutaneous coronary intervention in patients with double antiplatelet therapy: impact of genetic polymorphisms. Kreativnaya kardiologiya 2013(2), 15-27 (2013).
23. Mazurov AV, Ziuriaev IT, Khaspekova SG, Iakushkin VV, Sirotkina OV, Ruda M. Factors influencing platelet aggregation in patients with acute coronary syndrome. Terapevticheskii arkhiv 86(9), 83-89 (2014).
24. Mirzaev KB, Zelenskaya EM, Barbarash OL et al. CYP2C19 polymorphism frequency in Russian patients in Central Russia and Siberia with acute coronary syndrome. Pharmacogenomics and personalized medicine 10 107-114 (2017).
25. Galiavich AS, Valeeva DD, Minnetdinov R, Arkhipova AA, Akhmetov, Ii, Galiavi RA. CYP2C19 gene polymorphism in patients with myocardial infarction who use clopidogrel. Kardiologiia 52(4), 20-24 (2012).
26. Sumarokov AB, Meshkov AN, Buryachkovskaya LI et al. Polymorphism 416 Т>С OF gene CYP2C9 and sensitivity to clopidogrel. Tromboz, gemostazireologiya 2014(1), 53-61 (2014).
27. Beitelshees AL, Horenstein RB, Vesely MR, Mehra MR, Shuldiner AR. Pharmacogenetics and clopidogrel response in patients undergoing percutaneous coronary interventions. Clinical pharmacology and therapeutics 89(3), 455-459 (2011).
28. Bonello L, Armero S, Ait Mokhtar O et al. Clopidogrel loading dose adjustment according to platelet reactivity monitoring in patients carrying the 2C19*2 loss of function polymorphism. Journal of the American College of Cardiology 56(20), 1630-1636 (2010).
29. Collet JP, Hulot JS, Pena A et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet (London, England) 373(9660), 309-317 (2009).
30. Geisler T, Schaeffeler E, Dippon J et al. CYP2C19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics 9(9), 1251-1259 (2008).
31. Pedersen RS, Brasch-Andersen C, Sim SC et al. Linkage disequilibrium between the CYP2C19*17 allele and wildtype CYP2C8 and CYP2C9 alleles: identification of CYP2C haplotypes in healthy Nordic populations. European journal of clinical pharmacology 66(12), 1199-1205 (2010).
32. Sugimoto K, Uno T, Yamazaki H, Tateishi T. Limited frequency of the CYP2C19*17 allele and its minor role in a Japanese population. British journal of clinical pharmacology 65(3), 437-439 (2008).
33. Mirzaev KB, Sychev DA, Ryzhikova KA et al. Genetic Polymorphisms of Cytochrome P450 Enzymes and Transport Proteins in a Russian Population and Three Ethnic Groups of Dagestan. Genetic testing and molecular biomarkers 21(12), 747-753 (2017).
34. Feng GX, Liang Y, Bai Y et al. Clopidogrel metabolism related gene polymorphisms in Chinese patients with acute coronary syndrome. Zhonghua xin xue guan bing za zhi 40(11), 908-913 (2012).
35. Wang XQ, Shen CL, Wang BN, Huang XH, Hu ZL, Li J. Genetic polymorphisms of CYP2C19 2 and ABCB1 C3435T affect the pharmacokinetic and pharmacodynamic responses to clopidogrel in 401 patients with acute coronary syndrome. Gene 558(2), 200-207 (2015).
36. Khalikova AR, Arkhipova AA, Ahmetov II et al. The study of cytochrome P-450 CYP2C19 gene polymorphisms in population of Tatars living in Republic of Tatarstan. Prakticheskaja Medicina [Practical medicine] 2012(3), 53-55 (2012).
37. Kantemirova BI, Timofeeva NV, Sychev DA et al. A comparative study of CYP2C19 gene polymorphism in children living in the Astrakhanian region. . Astrahansk med zhurn 2011(3), 153-155 (2011).
38. Romodanovsky DP, Khapaev BA, Ignatiev IV et al. Frequencies of the «slow» allele variants of the genes coding isoenzymes of cytochrome Р450 CYP2D6, CYP2C19, CYP2C9 in Karachaevs and Circassians. Biomedicine 2010(2), 33-37 (2010).
39. Nizhevich AA, Yunusbaev BB, Tuygunov MM et al. Studying the polymorphisms of genes responsible for the metabolism of proton pump inhibitors in children with H. PYLORI infection: is there a connection with the effectiveness of eradication therapy? Experimental and clinical gastroenterology. 2009(3), 101-104 (2009).
40. Makeeva O, Stepanov V, Puzyrev V, Goldstein DB, Grossman I. Global pharmacogenetics: genetic substructure of Eurasian populations and its effect on variants of drug-metabolizing enzymes. Pharmacogenomics 9(7), 847-868 (2008).
41. Freidin MB, Bragina EYu, Petrouskiy FI et al. Association of the GSTT1, GSTM1, CYP2C19, CYP2E1 genes polymorphism with atopy. . Med. Immunol. 2003(5), 107-112 (2003).
42. Shuldiner AR, O’connell JR, Bliden KP et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. Jama 302(8), 849-857 (2009).
43. Frere C, Cuisset T, Morange PE et al. Effect of cytochrome p450 polymorphisms on platelet reactivity after treatment with clopidogrel in acute coronary syndrome. The American journal of cardiology 101(8), 1088-1093 (2008).
44. Giusti B, Gori AM, Marcucci R et al. Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4 IVS10 + 12G/A and P2Y12 T744C polymorphisms, is associated with response variability to dual antiplatelet treatment in high-risk vascular patients. Pharmacogenetics and genomics 17(12), 1057-1064 (2007).
45. Gurbel PA, Shuldiner AR, Bliden KP, Ryan K, Pakyz RE, Tantry US. The relation between CYP2C19 genotype and phenotype in stented patients on maintenance dual antiplatelet therapy. American heart journal 161(3), 598-604 (2011).
46. Price MJ, Murray SS, Angiolillo DJ et al. Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous coronary intervention: the GIFT (Genotype Information and Functional Testing) study. Journal of the American College of Cardiology 59(22), 1928-1937 (2012).
47. Hokimoto S, Mizobe M, Akasaka T et al. Impact of CYP2C19 polymorphism and proton pump inhibitors on platelet reactivity to clopidogrel and clinical outcomes following stent implantation. Thrombosis research 133(4), 599-605 (2014).
48. Tabata N, Hokimoto S, Akasaka T et al. Chronic kidney disease status modifies the association of CYP2C19 polymorphism in predicting clinical outcomes following coronary stent implantation. Thrombosis research 134(5), 939-944 (2014).
49. Frere C, Cuisset T, Gaborit B, Alessi MC, Hulot JS. The CYP2C19*17 allele is associated with better platelet response to clopidogrel in patients admitted for non-ST acute coronary syndrome. Journal of thrombosis and haemostasis : JTH 7(8), 1409-1411 (2009).
50. Tiroch KA, Sibbing D, Koch W et al. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. American heart journal 160(3), 506-512 (2010).
51. Cayla G, Hulot JS, O’connor SA et al. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. Jama 306(16), 1765-1774 (2011).
52. Sibbing D, Stegherr J, Latz W et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. European heart journal 30(8), 916-922 (2009).
53. Pare G, Mehta SR, Yusuf S et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. The New England journal of medicine 363(18), 1704-1714 (2010).
54. Hulot JS, Collet JP, Montalescot G. Genetic substudy of the PLATO trial. Lancet (London, England) 377(9766), 637, author reply 637-638 (2011).
55. Roberts JD, Wells GA, Le May MR et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet (London, England) 379(9827), 1705-1711 (2012).
56. So DY, Wells GA, Mcpherson R et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: the RAPID STEMI study. The pharmacogenomics journal 16(1), 71-78 (2016).
57. Holmes DR, Jr., Dehmer GJ, Kaul S, Leifer D, O’gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. Journal of the American College of Cardiology 56(4), 321-341 (2010).
58. Swen JJ, Nijenhuis M, De Boer A et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clinical pharmacology and therapeutics 89(5), 662-673 (2011).
59. Scott SA, Sangkuhl K, Stein CM et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clinical pharmacology and therapeutics 94(3), 317-323 (2013).
60. Ogawa H, Hokimoto S, Kaikita K et al. Current status and prospects of antiplatelet therapy in percutaneous coronary intervention in Japan: focus on adenosine diphosphate receptor inhibitors. Journal of cardiology 58(1), 6-17 (2011).
61. Price MJ, Berger PB, Teirstein PS et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. Jama 305(11), 1097-1105 (2011).
62. Trenk D, Stone GW, Gawaz M et al. A randomized trial of prasugrel versus clopidogrel in patients with high platelet reactivity on clopidogrel after elective percutaneous coronary intervention with implantation of drug-eluting stents: results of the TRIGGER-PCI (Testing Platelet Reactivity In Patients Undergoing Elective Stent Placement on Clopidogrel to Guide Alternative Therapy With Prasugrel) study. Journal of the American College of Cardiology 59(24), 2159-2164 (2012).
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Medical"
The word Medical refers to preventing or treating injuries or illnesses, relating to the study or practice of medicine. Medical care involves caring for a patient and helping them through their journey to recovery.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




