Structure and Binding Interface of Adeno-Associated Virus Serotype 9
Info: 6426 words (26 pages) Dissertation
Published: 18th May 2020
Tagged: Biology
Abstract
Adeno-associated viruses (AAVs) have been the focus of an increasing number of gene therapy research and clinical trials over the last two decades. Native AAVs are small, non-pathogenic parvoviruses that infect humans, integrating their viral DNA with the host DNA at a specific location on chromosome 19. Research utilizing AAVs as vectors for gene therapy has led to the discovery and development of multiple serotypes and genetic variants of these viruses towards a wide range of human disease models. The application of AAVs to neurological disorders is particularly appealing, as the low immunogenicity, consistent genomic delivery, and ability to infect both dividing and non-dividing cells gives AAVs a distinct advantage over current neurological gene therapy strategies. Last year, Luxturna – an AAV serotype 2 (AAV-2) therapeutic – was approved in the United States for the treatment of inherited retinal diseases. A second serotype, AAV-9, has demonstrated promising permeability through the blood brain barrier (BBB) and is being investigated in several studies for the treatment of neurological diseases through systematic delivery. One of the largest barriers to AAV efficacy in clinical trials thus far has been clearance of the therapeutic viruses by neutralizing antibodies in the patient. Recent studies have identified an antibody (ADK9) against AAV-9 that is involved in the neutralization of the virus. While the structure of AAV-9 alone has been solved via cryo-EM, as well as the structures of many other serotypes and serotype/antibody complexes, the structure of AAV-9 / ADK9 has not yet been elucidated. As of now, in vivo screening studies have implicated five residues in the immunogenicity of AAV-9 that are located in the same region as the binding interface of a related serotype/antibody complex, AAV-8 / ADK8. This study aims to characterize the interface of AAV-9 bound to ADK9 in order to elucidate the binding epitope of the complex and identify all contributing residues and their contacts. The identification of residues that participate in the complex interaction will be integral to the design of future AAV serotypes for more effective use in neurological disease therapies.
Specific Aim 1: Identify contributing antigenic residues in the AAV-9 / ADK9 interface
- Rational site-directed mutagenesis
- Analysis of epistasis between contributing residues
The goal of this aim is to identify the contribution from residues involved in the immunological interaction between AAV-9 and one of its naturally occurring monoclonal antibodies, ADK9. For the purposes of this aim, I will employ rational site-directed mutagenesis to analyze the effects on single, double, and triple mutations on the capsid/antibody complex.
Previous in vivo library screening has implicated residues 453-457 in the antigenicity of AAV-9 towards ADK9, so I will target these residues individually with mutations to alanine in order to investigate the contribution of each to the AAV-9 / ADK9 complex in the first round of experiments. All mutations will then be mixed in a combinatorial manner in order to collect further information on any epistasis in their contribution to antibody binding. The data collected within the scope of this aim will lay the groundwork for epitope mapping of the AAV-9 / ADK9 complex and can be used to design future mutagenesis experiments and refine structural data.
Specific Aim 2: Determine the structure of AAV-9 bound to ADK9
- Cryo-electron microscopy
The purpose of this aim is to solve the structure of AAV-9 bound to ADK9 using cryo-electron microscopy. Multiple existing cryo-EM and X-ray crystallography structures will be utilized as models for the success of this aim: that of AAV-9 alone, closely related serotype AAV-8 alone, the complex between AAV-8 and its antibody ADK8, and serotype AAV-5 with its antibody HL2476. Recent advances in cryo-electron microscopy have made the determination of high-resolution structures of these virus-antibody complexes possible. The data collected under the scope of this aim will be used to compliment previous methods for the determination of the epitope in the AAV-9 / ADK9 complex and will provide a foundation for further work in designing mutant AAV-9 capsids that have a higher level of evasion from the ADK9 antibody.
Background and Significance
Adeno-associated viruses (AAVs) are small, nonpathogenic viruses that infect and cause a minor immunological response in humans. Unlike pathogenic viruses, AAVs do not cause disease even in the presence of helper viruses, which are necessary for their replication (Naso, Tomkowicz, Iii, & Strohl, 2017; Wang, Tai, & Gao, 2019). They are generally cleared by neutralizing antibodies (NAbs) that bind to the surface proteins of the viral capsid and induce white blood cell mediated degradation (Bartel, Schaffer, & Büning, 2011; Tseng et al., 2016). If able to evade NAbs, the AAVs then bind to glycosylated cell surface receptors and enter the cell through clatherin-mediated endocytosis (Wang et al., 2019). The native form of this virus inserts its DNA into a conserved site on chromosome 19, termed the AAVS1 site (Kotin et al., 1990). In recent decades, gene therapy research has turned its attention to AAVs as a delivery system for genetic material. Recombinant AAVs (rAAVs) have been engineered so that the virus, lacking the wild-type viral protein genes, is no longer able to insert viral DNA into the host genome (Choi, McCarty, & Jude Samulski, 2006). Instead, inverted terminal repeats (ITRs) on either end of the desired transgenic DNA facilitate the formation of episomes that are able to survive in the nucleus of the infected cells (Choi et al., 2006). As the cells replicate, the transgenic DNA is diluted and cleared from the patient (Naso et al., 2017). This mechanism benefits the targeting of tissues with low cellular turnover, such as the central nervous system.
Adeno-associated virus structure
Since their accidental discovery in the 1960’s, the structure and biology of AAVs has been heavily studied (Atchison, Casto, & Hammon, 1965; Fields, Knipe, & Howley, 2007). Each non-envelope virus consists of a small (~26 nm diameter) icosahedral protein capsid containing a short single-stranded piece of DNA. Wild-type AAV DNA contains three genes (Cap / capsid, Rep / replication, and aap / assembly activating) flanked by ITRs and produces nine different gene products through translation modulation and splicing (Fields et al., 2007). The cap gene specifically codes for 3 viral proteins (VP1, VP2, and VP3) that form the capsid in a 1:1:10 ratio with 60 monomers total. All 3 viral proteins share an open reading frame and a C-terminus but differ in length at the N-terminus (Becerra, Koczot, Fabisch, & Rose, 1988). VP3 is the smallest of the three at 61 kDa, VP2 is extended partially to 73 kDa, and VP1 is the largest of the three at 87 kDa. Each viral protein has the same conserved parvovirus core structure: an eight-stranded beta barrel with an alpha helix.
Loops that connect the beta strands are sequentially and structurally variable between serotypes and are considered to be largely responsible for tissue tropism and antigenicity (Bell, Gurda, Van Vliet, Agbandje-Mckenna, & Wilson, 2012; DiMattia et al., 2012; Jose et al., 2018). The surface of the capsid has several distinguishing features due to these variable regions and the capsid geometry: there are rounded divots at the 5-fold axis, protrusions at the 3-fold axis, and depressions at the 2-fold axis next to raised areas termed the ‘2/5 walls’. Due to their contribution to antigenicity, tissue selectivity, and infection, these variable surface features are the focus of many in-depth structural studies (Gurda et al., 2013). Figure 1 illustrates the general structure of the viral capsid and the locations of the nine variable regions (VR-I – VR-IX).
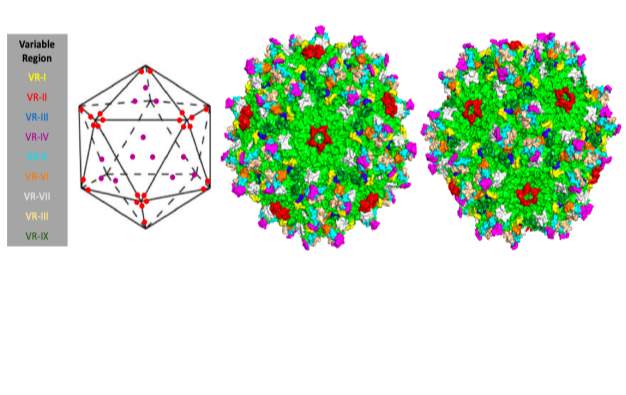
Variability between AAV serotypes
Several different serotypes of AAVs have been discovered, 13 of which have been identified in humans. Each individual serotype demonstrates a distinct tissue tropism, creating a natural focus on some serotypes over others for the treatment of specific diseases. Though the core structure of every AAV is the same, the amino acid sequence between serotypes can differ up to 50% (Daya & Berns, 2008). The nine variable regions have been delineated on the surface features of the viral capsid (Bell et al., 2012). The most studied serotype to date is recombinant AAV serotype 2 (rAAV-2), which was the first of the serotypes discovered and characterized. Multiple rAAV-2 drugs are currently being investigated in clinical trials, and two have already been approved (Glybera for the treatment of lipoprotein lipase deficiency in Europe and Luxturna for the treatment of inherited retina diseases in the United States) (FDA, 2017; Ylä-Herttuala, 2012). In recent years, the focus has expanded to lesser studied serotypes in order to achieve different specificities and evade immunological responses. Of the other identified human serotypes, AAV serotype 9 has demonstrated an elevated permeability across the blood brain barrier and is a promising alternative for the systematic delivery of therapeutics to the central nervous system (Foust et al., 2009).
Adeno-associated virus serotype 9
AAV serotype 9 is another naturally occurring serotype that was discovered in human liver tissue (Gao et al., 2004). The structure of AAV-9 has been determined to near-atomic resolution using both cryo-EM and X-ray crystallography (DiMattia et al., 2012). AAV-9 diverges from other human serotypes to different degrees in the variable regions on the surface structures of the viral particle. All nine variable regions are different between AAV-9 and AAV-4, for example, but only three regions differ between AAV-9 and AAV-2 and AAV-8 (DiMattia et al., 2012). AAV-9 has demonstrated increased delivery over other serotypes to numerous tissues, including cardiac, liver, skeletal muscle, and pancreas. Of particular interest is the unique ability of AAV-9 to cross the blood brain barrier and target the central nervous system with a higher efficiency than any other serotype identified so far, allowing for less invasive, systematic delivery options (Foust et al., 2009).
Immunological responses to AAV-9
Though adeno-associated viruses do not cause disease in humans, they elicit a minor immune response that clears the viral particles with neutralizing antibodies (Louis Jeune, Joergensen, Hajjar, & Weber, 2013). Clearance by NAbs has been identified as one of the largest barriers to therapeutic delivery of rAAVs (Naso et al., 2017; Tseng et al., 2016; Wang et al., 2019). The in vivo immunological response to AAV viruses is polyclonal, with multiple NAbs able to bind different epitopes on the surface of the viral capsid (Tseng et al., 2016). Current research suggests that most neutralizing antibodies interact with the variable regions on the viral proteins, reducing cross-reactivity as these areas contain the lowest degree of sequence homology between different serotypes (Gao et al., 2004; Gurda et al., 2013). Structural data on adeno-associated viruses and their antibodies, both separately and in a complex, have been an invaluable tool in the identification of residues involved in antigenicity. Recent studies have resulted in the solved the structures of all AAV serotypes in isolation, and the focus has begun to shift to AAV serotypes bound to monoclonal neutralizing antibodies (DiMattia et al., 2012; Jose et al., 2019).
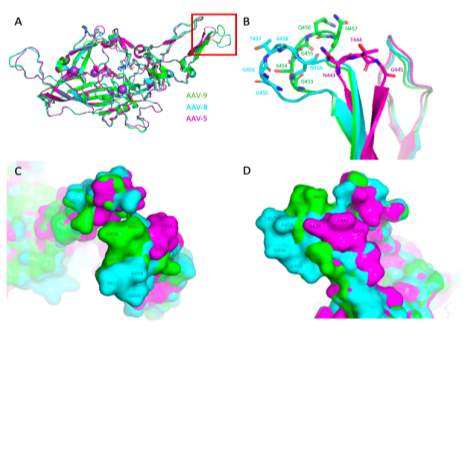
Until recently, only one anti-AAV-9 antibody (ADK9) had been identified (Sonntag et al., 2011). The binding surface and structure of AAV-9 bound to ADK9 remains unknown, though previous in vivo screening studies have identified AAV-9 VP residues 453-457, which are located in variable region IV on the 3-fold protrusion, as contributing to antigenicity towards ADK9 (Adachi, Enoki, Kawano, Veraz, & Nakai, 2014). A similar serotype, AAV-8 is bound in the same region by its monoclonal antibody ADK8 (Gurda et al., 2012). The structure and binding footprint of the AAV-8 / ADK8 complex was solved using cryo-EM and image reconstruction (Gurda et al., 2012). Recent advances in structure solution techniques for these viruses has resulted in the first atomic-level high-resolution cryo-EM structure of AAV-5 bound by one of its antibodies at the 3-fold protrusion (Jose et al., 2019). Both of these structures will be valuable tools during the structure solution of AAV-9 / ADK9 due to their similarity in sequence and binding location at the 3-fold protrusion. Figure 2 illustrates the structures of AAV-9, AAV-8, and AAV-5 overlaid. Compared to AAV-8, AAV-9 has smaller protrusions at the 3-fold axis, distinct topology at the raised 2/5 walls, and a small variation in the shape of the 2-fold depression – correlating to VR-IV, VR-I, and VR-II respectively (DiMattia et al., 2012). The VR-IV loop region of AAV-9 is slightly smaller than AAV-8, larger than AAV-5, and lies spatially between these two serotypes.
Significance
Adeno-associated viruses have quickly risen to the forefront of gene therapy research. Their low immunogenicity, broad infectious capabilities, tissue specificity, and low pathogenicity make them a powerful tool for the delivery of engineered genetic material. Currently, one of the largest barriers to rAAV success in the clinic is clearance by neutralizing antibodies. Among healthy adults, 30% – 80 % of individuals already have neutralizing antibodies against various AAV serotypes (Boutin et al., 2010; Calcedo, Vandenberghe, Gao, Lin, & Wilson, 2009). Right now, most rAAV gene therapy clinical trials require that patients be negative for all neutralizing antibodies in order to participate, barring a significant amount of people from potentially benefiting from these therapies. Improved understanding of the AAV-9 / ADK9 epitope and the specific interactions involved in recognition / binding will be instrumental to the design of evasive, therapeutic rAAV-9 molecules.
The detailed structural data that will be collected during the scope of this proposal will contribute to our understanding of the structure, serotype, and tissue tropism of adeno-associated virus 9, the only serotype that has demonstrated the ability to cross the blood brain barrier. This research will provide an important foundation for the design of future rAAV-9 viral capsids that can be delivered systematically, evade pre-existing neutralizing antibodies, and cross the BBB to deliver their genetic therapeutic.
Research Design
Specific Aim 1:
The overall goal of specific aim 1 is to characterize the contribution of AAV-9 residues 453-457 towards ADK9 binding. Rational site-directed mutagenesis coupled to a luciferase activity assay will be utilized to probe the network of interactions for each residue.
Rational Site-Directed Mutagenesis
All AAV-9 constructs will be expressed and purified using the recombinant AAV-9 viral-like particle plasmid in an insect-cell expression system described previously for this serotype (DiMattia et al., 2012; Mitchell et al., 2009; Tseng et al., 2016). Mutations will be made using the Quik Change II XL Site-Directed kit from Agilent.
1.1 Single Mutants
For the first round of mutagenesis experiments, each residue from 453-457 will be individually mutated to alanine. The positions of each of these residues in the wild-type AAV-9 viral capsid is shown in Figure 3. The ability of these mutant AAV-9 constructs to evade ADK9 will be compared to WT AAV-9 using luciferase neutralization assays performed in HeLa cells, as described by Tseng et al. (2016). In this assay, luciferase expression is measured in cells infected with the virus alone, in combination with ADK9, and in combination with ADK4. ADK4 is used in this assay as a negative control because of the very low degree of sequence similarity between AAV-4 and AAV-9 at the 3-fold protrusions and its previously demonstrated lack of AAV-9 clearance (Tseng et al., 2016). The viral capsid alone is used as a positive control for stability and the ability to infect cells in the absence of antibodies.
Figure 3 illustrates simulated data from WT and mutant AAV-9 constructs. In this example, the luciferase signal is attenuated to different degrees in the presence of the ADK9 antibody, which binds to the viral capsid and prevents it from infecting the HeLa cells. Some of the single
mutants, notably S454A and Q456A in this example, confer a significant increase in the ability of the capsid to evade ADK9, while mutants such as N457A do not demonstrate this effect. This data would implicate S454 and Q456 as frontrunners for further mutation studies that could optimize the efficacy of the viral capsid.
1.2 Double and Triple Mutant Epistasis
For the second round of mutagenesis experiments, a combinatorial mutation scheme will be performed to probe the epistasis between residues. For the five residues being studied here, there are ten different combinations of double mutants, and nine combinations of triple mutants. The same in vitro luciferase neutralization assay as described above will be performed on all double and triple mutants.
 Specific epistasis between combinations of residues occurs when the effect of two mutations is different than the sum of their individual contributions (Starr & Thornton, 2016). Positive epistasis with respect to ADK9 binding between two single mutations in the variable region of AAV-9 VP1 would
Specific epistasis between combinations of residues occurs when the effect of two mutations is different than the sum of their individual contributions (Starr & Thornton, 2016). Positive epistasis with respect to ADK9 binding between two single mutations in the variable region of AAV-9 VP1 would
indicate that they contribute,
in some way, to each other’s antigenicity. Negative epistasis would reflect the ability of a second mutation to ‘rescue’ the antibody binding lost by the first – in this case, not the desired outcome of the mutagenesis strategy. Measuring epistasis between different residues in the AAV-9 / ADK9 binding epitope will provide valuable information about the interactions between antigenic residues and will help inform the future rational design of more evasive AAV-9 vectors.
Examples of positive and negative epistasis from the data simulated above are illustrated in Figure 4. Multiple potential outcomes are represented among the ten double mutants. Epistasis between residues for the viral capsid alone gives insight into which residues may be important for cellular uptake and infection. Positive epistasis in these cases would indicate that the virus is able to infect and produce luciferase more efficiently, which might indicate a beneficial change in structure or stability of the VP regions that interact with the cell. With respect to ADK4 binding, positive epistasis would be an indication that binding had been ‘rescued’ by these mutations in combination with one another, creating undesirable cross-reactivity in the AAV-9 double mutant. Epistasis towards ADK9 binding gives a powerful insight into how AAV-9 residues are connected in their antigenicity. The Q456A mutation is of particular interest in the data set above, because it is involved in positive epistasis with three of the four other residues studied. This positive epistasis indicates that Q456A, in combination with many of its neighboring residues, increases AAV-9 evasion of ADK9. Such a result would inform further mutation studies at this position.
If this approach is not successful, an alternative method to quantifying viral uptake is centrifugation followed by quantitative PCR (qPCR). Instead of monitoring a signal from a reporter molecule expressed by infected cells, this method separates cells from free viral particles and then quantifies any transduced viral DNA. Such an approach could be used with various cells types and does not rely on the production of a recombinant gene product, which would provide the option to circumnavigate multiple steps in the luciferase neutralization assay described above if necessary.
Another alternative method would use surface plasmon resonance (SPR) to measure binding between the AAV-9 viral capsid and ADK9. In this method, the viral capsids are immobilized on a surface using amine coupling chemistry. The antibody in solution is applied briefly to the surface and then the system is regenerated. This process is repeated for multiple cycles, providing both binding affinity and binding kinetics data (Chenail, Brown, Shea, Feire, & Deng, 2011). This method would enable the quantitative analysis of the interaction between different AAV-9 constructs and ADK9 without relying on a cell-based assay.
Specific Aim 2:
 The goal of this aim is to gain structural information on the location and interactions of each residue that participates in the AAV-9 / ADK9 complex using cryo-electron microscopy. Improvements in optic equipment and modeling software have led to a renewed focus on cryo-EM for the determination of high (atomic-level) resolution protein structures (Cassidy, Himes, Luthey-Schulten, & Zhang, 2018; Serna, 2019). Recently, these advances have enabled the foot-printing and high resolution structures of different AAV serotypes bound to monoclonal antibodies (DiMattia et al., 2012; Gurda et al., 2012; Jose et al., 2019). These techniques will be combined with existing structural information to investigate the ADK9 epitope of AAV-9.
The goal of this aim is to gain structural information on the location and interactions of each residue that participates in the AAV-9 / ADK9 complex using cryo-electron microscopy. Improvements in optic equipment and modeling software have led to a renewed focus on cryo-EM for the determination of high (atomic-level) resolution protein structures (Cassidy, Himes, Luthey-Schulten, & Zhang, 2018; Serna, 2019). Recently, these advances have enabled the foot-printing and high resolution structures of different AAV serotypes bound to monoclonal antibodies (DiMattia et al., 2012; Gurda et al., 2012; Jose et al., 2019). These techniques will be combined with existing structural information to investigate the ADK9 epitope of AAV-9.
Cryo-electron microscopy to generate the structure of AAV-9 bound to ADK9
AAV-9 capsids will be expressed and purified using the recombinant AAV-9 viral-like particle plasmid in an insect-cell expression system described previously for this serotype (DiMattia et al., 2012; Mitchell et al., 2009; Tseng et al., 2016). Viral-like particles (VLPs) consist of the viral capsid without any genetic material. Only the Fab region of ADK9 will be used for these cryo-EM experiments. ADK9 Fabs are commonly generated using the cysteine protease papain on the full monoclonal antibody (Rader, 2009). The Fabs and VLPs will be mixed at a 120:1 ratio, resulting in a 2:1 ratio between each Fab and viral protein monomer (and thus, binding site) in the assembled capsid. The AAV-9 / ADK9 VLP/Fab complexes will be imaged using cryo-electron microscopy.
2.1 Cryo-electron microscopy
 The following protocol described by Jose et al. (2019) for the structure determination of AAV-5 bound to one of its monoclonal antibodies at the 3-fold protrusion will be utilized to achieve a high-resolution structure of AAV-9 / ADK9. Small amounts of AAV-9 / ADK9 will be added to glow-discharged copper EM grids with a 2-nm continuous carbon film, blotted, vitrified, and then analyzed for quality and particle distribution using images that will be collected on a Gatan UltraScan 4000 charge-coupled device (CCD) camera. Any grids that are deemed high enough quality for data collection will be used to collect micrographs using a Leginon semiautomated application on a Titan Krios electron microscope and will be recorded using a Gatan K2 summit direct electron detection camera. Data will be collected at a magnification of X130,000, which will give a pixel size of 1.07 angstroms. An example of such micrographs is shown in Figure 5.
The following protocol described by Jose et al. (2019) for the structure determination of AAV-5 bound to one of its monoclonal antibodies at the 3-fold protrusion will be utilized to achieve a high-resolution structure of AAV-9 / ADK9. Small amounts of AAV-9 / ADK9 will be added to glow-discharged copper EM grids with a 2-nm continuous carbon film, blotted, vitrified, and then analyzed for quality and particle distribution using images that will be collected on a Gatan UltraScan 4000 charge-coupled device (CCD) camera. Any grids that are deemed high enough quality for data collection will be used to collect micrographs using a Leginon semiautomated application on a Titan Krios electron microscope and will be recorded using a Gatan K2 summit direct electron detection camera. Data will be collected at a magnification of X130,000, which will give a pixel size of 1.07 angstroms. An example of such micrographs is shown in Figure 5.
2.2 Modeling the cryo-reconstruction of the AAV-9 / ADK9 complex
The structure of the AAV-9 VLP capsid bound to the ADK9 monoclonal antibody Fab will be modeled using several modeling programs and the AAV-8 / ADK8 and AAV-5 / HL2476 complexes as templates. AAV-8 / ADK8 was chosen as a template for this model because of the high sequence homology between AAV-8 and AAV-9 and the structural similarities at the 3-fold protrusion where ADK8 and ADK9 bind to the capsid (Gurda et al., 2012). AAV-5 / HL2476 was chosen as a template because of the extremely high resolution of the structure published earlier this year, as well as the binding location on the 3-fold protrusion (Jose et al., 2018).
First, the amino acid sequence of ADK9 light and heavy chains will be determined using cDNA from the hybridoma cell line. This sequence will be used to generate a model of the folded Fab for docking against AAV-9. The solved structure of AAV-9 (PDB code 3UX1) will be used for docking (DiMattia et al., 2012). The Rosetta docking software SnugDock will be used in tandem with SWISS-MODEL and PIGSpro to generate the complex model (Bordoli & Schwede, 2012; Guex, Peitsch, & Schwede, 2009; Lyskov et al., 2013; Lyskov & Gray, 2008; Sircar & Gray, 2010). The 3D model of the viral capsid bound to the ADK9 Fab will be used to analyze the density map obtained in 2.1. Finally, USCF Chimera will be employed to generate a final, atomic resolution structure from the micrograph data and all docked structural models.
An alternative approach to this aim is the use of chemical crosslinking mass spectrometry. In this method, AAV-9 is crosslinked to ADK9 using a partially hydrolyzed ion-labeled cross-linker in combination with Fourier transform ion-cyclotron resonance mass spectrometry (FTICR-MS) (Pimenova et al., 2008). This technique could be used as a separate avenue for structural determination, residue identification, and confirmation of previously published data and data collected from the mutagenesis experiments.
Abbreviations
|
AAV |
Adeno-associated virus |
|
Fab |
Antigen-binding fragment |
|
ITR |
Inverted terminal repeats |
|
NAbs |
Neutralizing antibodies |
|
rAAV |
Recombinant adeno-associated virus |
|
VLP |
Viral-like particle |
|
VR |
Variable Region (I – IX) |
Bibliography
- Adachi, K., Enoki, T., Kawano, Y., Veraz, M., & Nakai, H. (2014). Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nature Communications, 5. https://doi.org/10.1038/ncomms4075
- Atchison, R. W., Casto, B. C., & Hammon, W. M. (1965). Adenovirus-Associated Defective Virus Particles. Science, 149(3685), 754 LP – 755. https://doi.org/10.1126/science.149.3685.754
- Bartel, M., Schaffer, D., & Büning, H. (2011). Enhancing the clinical potential of aav vectors by capsid engineering to evade pre-existing immunity. Frontiers in Microbiology. Frontiers Research Foundation. https://doi.org/10.3389/fmicb.2011.00204
- Becerra, S. P., Koczot, F., Fabisch, P., & Rose, J. A. (1988). Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. Journal of Virology, 62(8), 2745–2754. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/2839699
- Bell, C. L., Gurda, B. L., Van Vliet, K., Agbandje-Mckenna, M., & Wilson, J. M. (2012). Identification of the Galactose Binding Domain of the Adeno-Associated Virus Serotype 9 Capsid. https://doi.org/10.1128/JVI.00448-12
- Bordoli, L., & Schwede, T. (2012). Automated protein structure modeling with swiss-model workspace and the protein model portal. Methods in Molecular Biology, 857, 107–136. https://doi.org/10.1007/978-1-61779-588-6_5
- Boutin, S., Monteilhet, V., Veron, P., Leborgne, C., Benveniste, O., Montus, M. F., & Masurier, C. (2010). Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Human Gene Therapy, 21(6), 704–712. https://doi.org/10.1089/hum.2009.182
- Calcedo, R., Vandenberghe, L. H., Gao, G., Lin, J., & Wilson, J. M. (2009). Worldwide Epidemiology of Neutralizing Antibodies to Adeno‐Associated Viruses. The Journal of Infectious Diseases, 199(3), 381–390. https://doi.org/10.1086/595830
- Cassidy, C. K., Himes, B. A., Luthey-Schulten, Z., & Zhang, P. (2018, June 1). CryoEM-based hybrid modeling approaches for structure determination. Current Opinion in Microbiology. Elsevier Ltd. https://doi.org/10.1016/j.mib.2017.10.002
- Cber, & Fda. (2017). December 19, 2017 Approval Letter – LUXTURNA. Retrieved from http://www.fda.gov/Drugs/DrugSafety/ucm400526.htm
- Chenail, G., Brown, N. E., Shea, A., Feire, A. L., & Deng, G. (2011). Real-time analysis of antibody interactions with whole enveloped human cytomegalovirus using surface plasmon resonance. Analytical Biochemistry, 411(1), 58–63. https://doi.org/10.1016/j.ab.2010.12.017
- Choi, V. W., McCarty, D. M., & Jude Samulski, R. (2006). Host Cell DNA Repair Pathways in Adeno-Associated Viral Genome Processing. JOURNAL OF VIROLOGY, 80(21), 10346–10356. https://doi.org/10.1128/JVI.00841-06
- Daya, S., & Berns, K. I. (2008). Gene Therapy Using Adeno-Associated Virus Vectors. Clinical Microbiology Reviews, 21(4), 583–593. https://doi.org/10.1128/CMR.00008-08
- DiMattia, M. A., Nam, H.-J., Van Vliet, K., Mitchell, M., Bennett, A., Gurda, B. L., … Agbandje-McKenna, M. (2012). Structural Insight into the Unique Properties of Adeno-Associated Virus Serotype 9. Journal of Virology, 86(12), 6947–6958. https://doi.org/10.1128/jvi.07232-11
- Fields, B. N., Knipe, D. M., & Howley, P. M. (2007). Fields virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins.
- Foust, K. D., Nurre, E., Montgomery, C. L., Hernandez, A., Chan, C. M., & Kaspar, B. K. (2009). Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nature Biotechnology, 27(1), 59–65. https://doi.org/10.1038/nbt.1515
- Gao, G., Vandenberghe, L. H., Alvira, M. R., Lu, Y., Calcedo, R., Zhou, X., & Wilson, J. M. (2004). Clades of Adeno-associated viruses are widely disseminated in human tissues. Journal of Virology, 78(12), 6381–6388. https://doi.org/10.1128/JVI.78.12.6381-6388.2004
- Guex, N., Peitsch, M. C., & Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis, 30(SUPPL. 1). https://doi.org/10.1002/elps.200900140
- Gurda, B. L., DiMattia, M. A., Miller, E. B., Bennett, A., McKenna, R., Weichert, W. S., … Agbandje-McKenna, M. (2013). Capsid Antibodies to Different Adeno-Associated Virus Serotypes Bind Common Regions. Journal of Virology, 87(16), 9111–9124. https://doi.org/10.1128/jvi.00622-13
- Gurda, B. L., Raupp, C., Popa-Wagner, R., Naumer, M., Olson, N. H., Ng, R., … Agbandje-McKenna, M. (2012). Mapping a Neutralizing Epitope onto the Capsid of Adeno-Associated Virus Serotype 8. Journal of Virology, 86(15), 7739–7751. https://doi.org/10.1128/jvi.00218-12
- Jose, A., Mietzsch, M., Smith, J. K., Kurian, J., Chipman, P., McKenna, R., … Agbandje-McKenna, M. (2018). High-Resolution Structural Characterization of a New Adeno-associated Virus Serotype 5 Antibody Epitope toward Engineering Antibody-Resistant Recombinant Gene Delivery Vectors. Journal of Virology, 93(1), 1–16. https://doi.org/10.1128/jvi.01394-18
- Jose, A., Mietzsch, M., Smith, J. K., Kurian, J., Chipman, P., Mckenna, R., … Sandri-Goldin, R. M. (2019). High-Resolution Structural Characterization of a New Adeno-associated Virus Serotype 5 Antibody Epitope toward Engineering Antibody-Resistant Recombinant Gene Delivery Vectors. Jvi.Asm.Org 1 Journal of Virology, 93, 1394–1412. https://doi.org/10.1128/JVI.01394-18
- Kotin, R. M., Siniscalco, M., Samulski, R. J., Zhu, X., Hunter, L. A., Laughlin, C. A., … Berns, K. I. (1990). Site-specific integration by adeno-associated virus. Proceedings of the National Academy of Sciences, 87(6), 2211–2215. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=53656&tool=pmcentrez&rendertype=abstract
- Louis Jeune, V., Joergensen, J. A., Hajjar, R. J., & Weber, T. (2013, April 1). Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Human Gene Therapy Methods. https://doi.org/10.1089/hgtb.2012.243
- Lyskov, S., Chou, F. C., Conchúir, S. Ó., Der, B. S., Drew, K., Kuroda, D., … Das, R. (2013). Serverification of Molecular Modeling Applications: The Rosetta Online Server That Includes Everyone (ROSIE). PLoS ONE, 8(5). https://doi.org/10.1371/journal.pone.0063906
- Lyskov, S., & Gray, J. J. (2008). The RosettaDock server for local protein-protein docking. Nucleic Acids Research, 36(Web Server issue). https://doi.org/10.1093/nar/gkn216
- Mitchell, M., Nam, H. J., Carter, A., McCall, A., Rence, C., Bennett, A., … Agbandje-Mckenna, M. (2009). Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 9. Acta Crystallographica Section F: Structural Biology and Crystallization Communications, 65(7), 715–718. https://doi.org/10.1107/S1744309109021460
- Naso, M. F., Tomkowicz, B., Iii, W. L. P., & Strohl, W. R. (2017). Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs, 31. https://doi.org/10.1007/s40259-017-0234-5
- Pimenova, T., Nazabal, A., Roschitzki, B., Seebacher, J., Rinner, O., & Zenobi, R. (2008). Epitope mapping on bovine prion protein using chemical cross-linking and mass spectrometry. JOURNAL OF MASS SPECTROMETRY J. Mass Spectrom, 43, 185–195. https://doi.org/10.1002/jms.1280
- Rader, C. (2009). Overview on concepts and applications of fab antibody fragments. Current Protocols in Protein Science. https://doi.org/10.1002/0471140864.ps0609s55
- Serna, M. (2019). Hands on methods for high resolution cryo-electron microscopy structures of heterogeneous macromolecular complexes. Frontiers in Molecular Biosciences. Frontiers Media S.A. https://doi.org/10.3389/fmolb.2019.00033
- Sircar, A., & Gray, J. J. (2010). SnugDock: Paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Computational Biology, 6(1). https://doi.org/10.1371/journal.pcbi.1000644
- Sonntag, F., Köther, † K, Schmidt, ‡ K, Weghofer, M., Raupp, C., Nieto, K., … Kleinschmidt, J. A. (2011). The Assembly-Activating Protein Promotes Capsid Assembly of Different Adeno-Associated Virus Serotypes. Erwin-Rentschler Strasse, 85(23), 12686–12697. https://doi.org/10.1128/JVI.05359-11
- Starr, T. N., & Thornton, J. W. (2016, July 1). Epistasis in protein evolution. Protein Science. Blackwell Publishing Ltd. https://doi.org/10.1002/pro.2897
- Tseng, Y. S., Vliet, K. Van, Rao, L., McKenna, R., Byrne, B. J., Asokan, A., & Agbandje-McKenna, M. (2016). Generation and characterization of anti-Adeno-associated virus serotype 8 (AAV8) and anti-AAV9 monoclonal antibodies. Journal of Virological Methods, 236, 105–110. https://doi.org/10.1016/j.jviromet.2016.07.009
- Wang, D., Tai, P. W. L., & Gao, G. (2019). Adeno-associated virus vector as a platform for gene therapy delivery. Nature Reviews Drug Discovery, 18(5), 358–378. https://doi.org/10.1038/s41573-019-0012-9
- Ylä-Herttuala, S. (2012). Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Molecular Therapy : The Journal of the American Society of Gene Therapy, 20(10), 1831–1832. https://doi.org/10.1038/mt.2012.194
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Biology"
Biology is the scientific study of the natural processes of living organisms or life in all its forms. including origin, growth, reproduction, structure, and behaviour and encompasses numerous fields such as botany, zoology, mycology, and microbiology.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




